Abstract
The complete mitochondrial genome sequence of Budorcas taxicolor bedfordi was 16 662-bp long, containing 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes and a control region. All of the PCGs begin with the typical ATN start codon. Stop codons TAA and AGA are present in the PCGs; exceptions are ND2, COIII, ND3 and ND4, which possess incomplete termination codon (T––). Secondary structure prediction of the 22 tRNA genes revealed the absence of the DHU arm in tRNASer(AGN).
Golden takin, Budorcas taxicolor bedfordi, is uniquely distributed in Qinling Mountains in China. It is classified as Vulnerable (VU) under International Union for the Conservation of Nature and Natural Resources (IUCN) (2008) criteria, and is a first-class national protected animal in China. The limited molecular data dampen the phylogenetic and evolution studies on B. t. bedfordi. Here, the complete mitochondrial genome of B. t. bedfordi has been reported with the general features. An illegal hunting B. t. bedfordi was collected by forest police in Qinling Mountains (33°47′N′, 107°42′E), Shaanxi, China. This specimen is stored in Shaanxi Key Laboratory for Animal Conservation (accession number LN13112). The mitochondrial genome was determined after PCR amplification, sequencing and annotation.
The complete mitochondrial genome of B. t. bedfordi is 16 662 bp in length and has been deposited in GenBank (accession no. KU361169). Its genes arrangement, order and orientation are identical to that of typical mammal mtDNA (Shen et al. Citation2012). There are 13 intergenic spacer regions, ranging in size from 1 to 7 bp (21 bp in total). Gene overlap involves 72 bp over seven locations. ATP8 and ATP6 have the longest overlap (40 bp). The overall base composition of the whole mitochondrial genome was A (33.9%), T (27.0%), C (26.3%) and G (12.8%), with an AT bias of 60.9%. The typical ATN (ATG or ATT) start codon is present in all of the 13 B. t. bedfordi PCGs. Open-reading frames of most B. t. bedfordi genes end with TAA or AGA, while ND2, COIII, ND3 and ND4 have the incomplete stop codon T––. All the tRNA genes that can fold into cloverleaf secondary structures except for the shortest gene tRNASer(AGY) (60 bp), in which the DHU arm has been replaced by a simple loop.
The A + T-rich region, which is located between srRNA and tRNAIle, is 1233 bp in length. There were 2 repeats of a short core sequence of alternating purines and pyrimidines and constituting a GYRCAT (Y = C/T, R = A/G) motif in the RS2 region of ETAS region. F, E, D, C, B-box, and CSB1-box elements (Douzery & Randi Citation1997; E Sbisa et al. Citation1997; Guo et al. Citation2015; Iyengar et al. Citation2006).
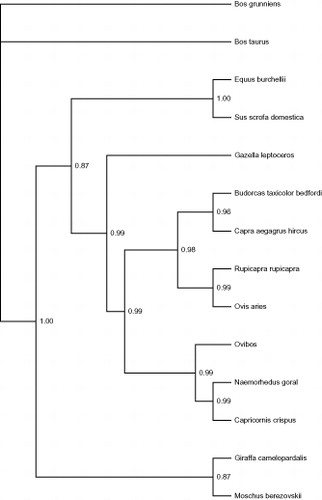
To further validate the mitogenome of B. t. bedfordi and research the phylogenetic relationships of Bovidae, the phylogenetic analyses were performed using MrBayes 3.1.2 (Ronquist & Huelsenbeck Citation2003) based on the whole mitogenome of B. t. bedfordi and the other 13 taxa that were retrieved from GenBank (KR011113, AF492351, KR059225, AP003429, JX312729, NC020708, AP003424, NC012694, NC021381, KF826487, NC001941, NC020633, AP003428) ().
Funding information
The study was supported by Science and Technology Foundation of Shaanxi Academy of Sciences, China (2009K-02, 2013K-04), Natural Science Foundation of Shaanxi Province, China (2013JM3016, 2015NY060).
Disclosure statement
The authors declare no competing interests.
References
- Douzery E, Randi E. 1997. The mitochondrial control region of cervidae: evolutionary patterns and phylogenetic content. Mol Biol Evol. 14:1154–1166.
- Guo X, Pei J, Bao PJ, Chu M, Wu XY, Ding XZ, Yan P. 2015. The complete mitochondrial genome of the Qinghai Plateau yak Bos grunniens (Cetartiodactyla: Bovidae). Mitochondrial DNA. [Online] [cited 2015 Oct 20]. Available at: http://www.tandfonline.com/doi/full/10.3109/19401736.2015.1060423.
- Iyengar A, Diniz FM, Gilbert CT, Woodfine T, Knowles J, Maclean N. 2006. Structure and evolution of the mitochondrial control region in oryx. Mol Phylogen Evol. 40:305–314.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Sbisà E, Tanzariello F, Reyes A, Pesole G, Saccone C. 1997. Mammalian mitochondrial D-loop region structural analysis: identification of new conserved sequences and their functional and evolutionary implications. Gene. 205:125–140.
- Shen X, Tian M, Meng XP, Liu HL, Cheng HL, Zhu CB, Zhao FQ. 2012. Complete mitochondrial genome of Membranipora grandicella (Bryozoa: Cheilostomatida) determined with next-generation sequencing: the first representative of the suborder Malacostegina. Comparative Biochem Physiol Part D 7:248–253.