Abstract
The complete mitochondrial genome of Yaoshania pachychilus (Protomyzon pachychilus) was determined for the first time. This mitogenome was 16 564 bp in length, including two ribosomal RNA genes, 22 transfer RNA genes, containing 13 protein-coding genes and a control region (D-loop). The base composition of the heavy strand was 28.9% A, 25.2% T, 28.7% C and 17.2% G. As in other vertebrates, with the exception of the eight tRNA genes and NADH dehydrogenase subunit 6 (ND6), all other mitochondrial genes are encoded on the heavy strand. Only the tRNA-Ser was not folded into a typical clover-leaf secondary structure as it lacks the dihydrouridine arm.
Yaoshania pachychilus (Teleostei, Cypriniformes, Homalopteridae, Protomyzon) is a rare fish found in China belonging to the family of Homalopteridae which has two subfamilies, a total of about 37 genera, 470 species (Yang et al. Citation2012). Yaoshania pachychilus is a demersal freshwater fish and it lives on hill rapids, the high mountain lakes and rivers. Yaoshania pachychilus is a small ornamental fish belonging to cold fish, and it has been listed in China red data book of endangered animals.
In this study, we have presented the complete mitochondrial genome of Yaoshania pachychilus. The specimens were collected from Dayaoshan Mountain, Jinxiu County, Xi Jiang, geographical coordinates: longitude 113°11′–113°16′, north latitude 25°07′–25°14′ of Guangxi Province, China. The primary specimens had been stored in herbarium in Kunming Institute of Zoology (KIZ), Chinese Academy of Sciences and the accession number is 200243055. This information can be queried in the National Specimen Information Infrastructure (NSII) of China. The complete mitogenome of Yaoshania pachychilus was 16 564 bp in size with 13 protein-coding genes, 22 tRNA genes and a control region (GenBank accession No. KT031050). Most of the mitochondrial genes were encoded on heavy strand except for NADH dehydrogenase subunit 6 (ND6) and eight tRNA genes (tRNA-Gln, -Ala, -Asn, -Cys, -Tyr, -Ser1, -Glu and -Pro) which were encoded on light strand (Boore Citation1999). The gene composition, arrangement and transcriptional orientation in Yaoshania pachychilus are similar to other vertebrates (). The overall base composition of the H-strand was 28.9% A, 25.2% T, 28.7% C and 17.2% G. All protein-coding genes started with the standard ATG codon except COX1 gene, which used GTG as the starting codon. TAA was the typical stop codon in seven protein-encoding genes, while TAG was the stop codon for protein-coding genes ND1, ND2, ND3 and ND4, and other two genes (COII and Cytb) had incomplete stop codon T. The 12S rRNA (952 bp) and 16S rRNA (1678 bp) genes were separated by the tRNA-Val gene. Twenty-two tRNA genes ranged from 66 bp (tRNA-Cys) to 76 bp (tRNA-Gly1). Only tRNA-Ser2 lacked the dihydrouridine arm and replaced it with a simple loop, the remaining tRNAs could be folded into a typical clover-leaf secondary structure (Liang et al. Citation2014). Between tRNA-Asn and tRNA-Cys, a 31-bp sequence was identified as the origin of L-strand replication (OL), which could be folded into a hairpin structure (8 bp stem and 13 bp loop). The A + T-rich (63.31%) control region (905 bp) was located between tRNA-Pro and tRNA-Phe. The termination-associated sequence (TAS) and the conserved sequence blocks (CSB1-3) were found in the control region.
Table 1. Characteristics of the mitochondrial genome of Yaoshania pachychilus
We had proceeded the molecular evolution analysis of mitochondrial DNA by using MEGA5.1 (Tamura et al. Citation2011). Phylogenetic analysis indicated that Yaoshania pachychilus had the closest relationship with Metahomaloptera omeiensis (Teleostei, Cypriniformes, Homalopteridae, Metahomalopterinae) (Li et al. Citation2014) which is a demersal freshwater fish ().
Figure. 1. Phylogenetic tree was constructed using whole mitogenomes of Yaoshania pachychilus and other closely related organisms.
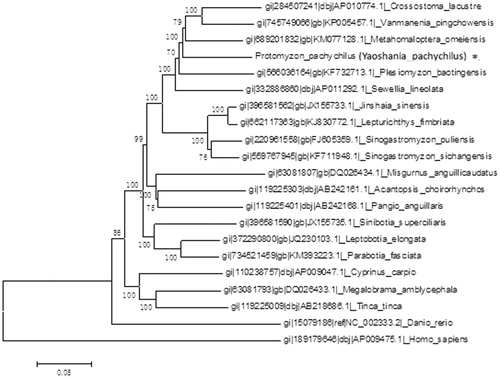
We think these data will contribute to uncover the related generic groups, the genetic and evolutionary relationships among Homalopteridae in the Cypriniformes in different areas of Chinese Mainland and Asia.
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for doing the research and writing the paper.
Funding information
This work was supported by the Graduate Student Research Innovation Project in Hunan Province in China [grant number CX2014B201].
References
- Yang J, Kottelat M, Yang JX, Chen XY. 2012. Yaoshania and Erromyzon kalotaenia, a new genus and a new species of balitorid loaches from Guangxi, China (Teleostei: Cypriniformes). Zootaxa. 3586:173–186.
- Tang WJ, Chen YY. 2000. Study on taxonomy of Homalopteridae. J Shanghai Fish Univ. 9:1–10 (in Chinese with English abstract).
- Li HJ, Zhao HX, Lin XT, Chen M. 2010. Fish communities in Dayaoshan State Natural Reserve of Guangxi, China. J Henan Normal Univ (Nat Sci). 38:123–127 (in Chinese with English abstract).
- Chen M, Huang N, Li HJ. 2002. Study on the fishes of Homalopteridae and their distribution from Guangxi. J Xinyang Teach Coll (Nat Sci Ed). 15:204–207 (in Chinese with English abstract).
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Liang Z, Wang C, Wu Y, Li H, Yuan X, Wei Q. 2014. Complete mitochondrial genome of Vanmanenia pingchowensis (Cypriniformes, Cyprinidae). Mitochondrial DNA. 27:1–2.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 28:2731–2739.
- Li Y, Tang M, Xue Y, Chen HJ, Ye Q. 2014. Complete mitochondrial genome of the Chinese balitorine loach, Metahomaloptera omeiensis (Teleostei, Cypriniformes). Mitochondrial DNA. 27:1449–1450.
