Abstract
The spotted-wing drosophila, Drosophila suzukii (Diptera: Drosophilidae) is an Asian species introduced into North America and Europe. It damages a wide variety of thin-skinned fruits. We sequenced the complete mitochondrial genome (mitogenome) to better understand the mitogenomic characteristics of this species. The 16 230-bp complete mitogenome of the species consists of a typical set of genes, including 13 protein-coding genes (PCGs), two rRNA genes and 22 tRNA genes, and one major non-coding A + T-rich region, with an arrangement typical of insects. Twelve PCGs began with the typical ATN codon, whereas the COI began with TCG, which has been designated as the start codon for other Drosophila species. The 1525-bp A + T-rich region is the second longest in Drosophila species for which the whole mitogenome has been sequenced, after D. melanogaster. Phylogenetic analysis using the 13 PCGs of the Drosophila species indicated that D. suzukii is placed, with a strong support, in the basal lineage of the previously defined Melanogaster group.
Drosophila suzukii (Diptera: Drosophilidae) is an economically damaging pest. The females have serrated ovipositor, which enables them to pierce most thin-skinned fruits (Walsh et al. Citation2011). D. suzukii is an Asian species that has been recorded in China, India, Japan, South Korea and Thailand (Hauser Citation2011), but has invaded the United States, British Columbia, Italy, France and Spain (Toda Citation1991; Hauser et al. Citation2009; Lee et al. Citation2011).
In this study, we sequenced the complete mitochondrial genome (mitogenome) of D. suzukii to better understand the mitogenomic characteristics of this species and its phylogenetic relationships within Drosophila. One adult was captured from an arboretum located in Gwangju City, South Korea (35°10′21.6″ N, 126°53′57.6″ E). A voucher specimen was deposited in Chonnam National University, Gwangju, Korea.
By using the total DNA as template, two long overlapping fragments (COI–CytB and CytB–COI) were amplified, and subsequently, 26 short overlapping fragments were amplified using the two long fragments as templates. Primers for the long and short fragments were designed in this study by using available Drosophila mitogenome sequences (Andrianov et al. Citation2010; De Ré et al. Citation2014; Llopart et al. Citation2014).
The D. suzukii mitogenome is 16 230 bp in length and includes the typical sets of genes (two rRNAs, 22 tRNAs and 13 protein-coding genes [PCGs]) and a major non-coding A + T-rich region (GenBank accession number: KU588141). The mitogenome size is well within the range found in Drosophila, from 15 450 bp (D. incompta; De Ré et al. Citation2014) to 19 317 bp (D. melanogaster; Unpublished, GenBank accession number: KJ947872). The 1525-bp A + T-rich region is the second longest among sequenced Drosophila, after the 4608-bp A + T-rich region of D. melanogaster. However, despite the length, the D. suzukii A + T-rich region does not have long tandem repeats. Instead, it has interspersed TA repeat, poly-T stretch and poly-A stretch. The gene arrangement of D. suzukii is identical to that of the ancestral insect order found in majority of insects (Boore Citation1999).
The AT content among genes and regions varies profoundly in the D. suzukii mitogenome, including the A + T-rich region (92.5%), lrRNA (82.7%), srRNA (78.9%), trns (75.9%) and PCGs (76.23%). Twelve D. suzukii PCGs began with the typical ATN codon (six with ATG, four with ATT, one with ATC and one with ATA), whereas COI began with the atypical sequence TCG (Serine), as has been proposed in other species such as D. littoralis, D. pseudoobscura, D. santomea, D. yakuba and D. melanogaster (Clary & Wolstenholme Citation1985; Torres et al. Citation2009; Andrianov et al. Citation2010; Llopart et al. Citation2014; Unpublished, GenBank accession number: KJ947872).
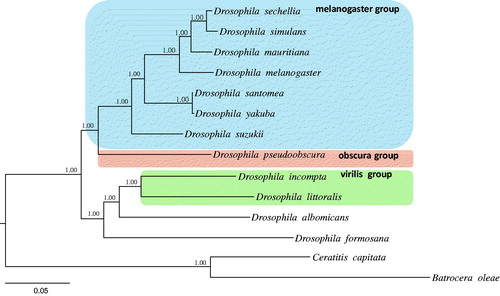
We performed a phylogenetic analysis by using the 13 PCGs and the available mitogenome sequences of 11 Drosophila downloaded from GenBank and two species of Tephritidae as outgroups. Bayesian inference method was performed using the GTR + GAMMA + I model in CIPRES Portal v. 3.1 (Miller et al. Citation2010). The results showed that D. suzukii is placed as the basal lineage of the previously defined Melanogaster group (Da Lage et al. Citation2007), with a strong support (Bayesian posterior probabilities = 1.0). The Melanogaster group included the following species: D. sechellia, D. simulans, D. mauritiana, D. melanogaster, D. santomea and D. yakuba ().
Figure 1. Phylogeny of Drosophila constructed using the Bayesian inference method by analyzing 13 protein-coding genes (PCGs). Values at each node specify Bayesian posterior probabilities in percent. The scale bar indicates number of substitutions per site. Two species of Tephritidae were included as outgroups. GenBank accession numbers are as follows: D. suzukii, KU588141; D. albomicans, KT119344; D. incompta, KM275233; D. littoralis, FJ447340; D. pseudoobscura, FJ899745; D. mauritiana, AF200830; D. santomea, KF824856; D. sechellia, AF200832; D. simulans, AF200833; D. yakuba, NC_001322; D. melanogaster, KJ947872; D. formosana, KR265324; Ceratitis capitate, NC_000857.1 and Bactrocera oleae, NC_005333.1.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding information
This research was supported by a grant (22143383400) from the Animal and Plant Quarantine Agency, South Korea.
References
- Andrianov B, Goryacheva I, Sorokina S, Gorelova T, Mitrofanov V. 2010. Comparative analysis of the mitochondrial genomes in Drosophila virilis species group (Diptera: Drosophilidae). Trends Evol Biol. 2:e4.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22:252–271.
- Da Lage JL, Kergoat GJ, Maczkowiak F, Silvain JF, Cariou ML, Lachaise D. 2007. A phylogeny of Drosophilidae using the Amyrel gene: questioning the Drosophila melanogaster species group boundaries. J Zool Syst Evol Res. 45:47–63.
- De Ré FC, Wallau GL, Robe LJ, Loreto ELS. 2014. Characterization of the complete mitochondrial genome of flower-breeding Drosophila incompta (Diptera, Drosophilidae). Genetica. 142:525–535.
- Hauser M. 2011. A historic account of the invasion of Drosophila suzukii (Matsumura) (Diptera: Drosophilidae) in the continental United States, with remarks on their identification. Pest Manag Sci. 67:1352–1357.
- Hauser M, Gaimari S, Damus M. 2009. Drosophila suzukii new to North America. Fly Times. 43:12–15.
- Lee JC, Bruck DJ, Dreves AJ, Ioriatti C, Vogt H, Baufeld P. 2011. In focus: spotted wing drosophila, Drosophila suzukii, across perspectives. Pest Manag Sci. 67:1349–1351.
- Llopart, A, Herrig, D, Brud, E, Stecklein, Z. 2014. Sequential adaptive introgression of the mitochondrial genome in Drosophila yakuba and Drosophila santomea. Mol Ecol. 23:1124–1136.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proc Gateway Comput Environ Workshop (GCE), New Orleans, pp. 1–8.
- Toda MJ. 1991. Drosophilidae (Diptera) in Myanmar (Burma) VII. The Drosophila melanogaster species-group, excepting the D. montium species sub-group. Orient Insects. 25:69–94.
- Torres TT, Dolezal M, Schlotterer C, Ottenwalder B. 2009. Expression profiling of Drosophila mitochondrial genes via deep mRNA sequencing. Nucleic Acids Res. 37:7509–7518.
- Walsh DB, Bolda MP, Goodhue RE, Dreves AJ, Lee JC, Bruck DJ, Walton VM, O’Neal SD, Zalom FG. 2011. Drosophila suzukii (Diptera: Drosophilidae): invasive pest of ripening soft fruit expanding its geographic range and damage potential. J Integr Pest Manag. 1:1–7.
