Abstract
Taraxacum officinale is a distributed weedy plant used as a traditional medicinal herb belonging to the family Asteraceae. The complete chloroplast genome of T. officinale was generated by de novo assembly with whole genome sequence data. The chloroplast genome was 151 324 bp in length, which consisted of a large single copy region of 83 895 bp and a short single copy region of 18 549 bp separated by a pair of inverted repeat regions of 24 440 bp. The chloroplast genome contained 79 protein-coding genes, 29 tRNA genes and four rRNA genes. Phylogenetic analysis revealed that T. officinale was closely related to Lactuca sativa.
The genus Taraxacum, known as dandelion, belongs to the family Asteraceae and widely distributed in the temperate areas of the Northern Hemisphere. More than 2500 species are known in the genus Taraxacum (Richards Citation1973; Ge et al. Citation2007). Taraxacum species have long been utilized for medicinal purposes due to their various pharmacological activities including diuretic and anti-inflammatory effects. Taraxacum roots include various sesquiterpens, triterpenes, phytosterols, flavonoids and phenolic compounds (Schütz et al. Citation2006). The genus Taraxacum shows taxonomic complexity due to the presence of agamospermy, complex hybrids and polyploidy, which makes taxonomic studies of this genus difficult (Ge et al. Citation2007). So far, complete chloroplast genome sequences of many species in the family Asteraceae have been reported including Helianthus annuus and Artemisia frigida (Timme et al. Citation2007; Liu et al. Citation2013), whereas none of Taraxacum species was subject to study on complete chloroplast genome sequence. On this account, we generated the complete chloroplast genome sequence of T. officinale for the first time in the genus Taraxacum. The sequence will provide the information for phylogenetic analysis and molecular study of this species as well as others in the family Asteraceae.
Total genomic DNA was extracted from leaves of T. officinale collected from Hantaek Botanical Garden (http://www.hantaek.co.kr), Yongin, Korea, using a modified cetyltrimethylammonium bromide protocol (Allen et al. Citation2006). A paired-end (PE) library with 300-bp insert size was constructed and sequenced using an Illumina MiSeq instrument (Illumina, San Diego, CA) by Lab Genomics Co. (http://www.labgenomics.co.kr), Seoul, Korea. Whole genome sequence data of 1.3 Gb were generated, trimmed, and assembled by a CLC genome assembler (v. beta 4.6, CLC Inc., Aarhus, Denmark), as described in Kim et al. (Citation2015a,Citationb). The representative chloroplast contigs were retrieved, ordered and combined into a single draft sequence, through comparison with the chloroplast sequence of Panax ginseng (GenBank accession no. NC_006290) as a guidance. The draft sequence was validated and manually corrected by PE read mapping. The protein-coding, tRNA and rRNA genes in the chloroplast genome were annotated using the DOGMA program (Wyman et al. Citation2004) and manual curation based on BLAST searches.
The complete chloroplast genome of T. officinale (GenBank accession no. KU361241) was 151 324 bp in length and showed a typical quadripartite chloroplast genome structure consisting of a large single copy region of 83 895 bp, a short single copy region of 18 549 bp, and a pair of inverted repeats regions of 24 440 bp. The chloroplast genome contained a total of 112 genes including 79 protein-coding genes, 29 tRNA genes and 4 rRNA genes. The GC content of the whole chloroplast genome was 37.7%.
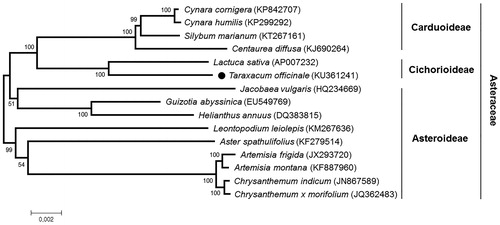
Phylogenetic analysis was conducted with common 68 chloroplast protein coding sequences of T. officinale and other 14 species belonging to the family Asteraceae, using a maximum likelihood analysis of MEGA 6.0 (Tamura et al. Citation2013) with 1000 bootstrap replicates. The phylogenetic tree indicated that T. officinale was most closely related to Lactuca sativa belonging to the subfamily Cichorioideae, as expected ().
Disclosure statement
The authors report that they have no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This work was supported by “15172MFDS246” from Ministry of Food and Drug Safety in 2015, Republic of Korea.
References
- Allen G, Flores-Vergara M, Krasynanski S, Kumar S, Thompson W. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1:2320–2325.
- Ge X, Kirschner J, Štěpánek J. 2007. Taraxacum. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China, vols. 20–21. Beijing: Science Press/St. Louis MO: Botanical Garden Press; [cited 2016 Jan 20]. Available at: http://www.efloras.org/florataxon.aspx?flora_id=2&taxon_id=132314.
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ. 2015a. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 10:e0117159.
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015b. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Liu Y, Huo N, Dong L, Wang Y, Zhang S, Young HA, Feng X, Gu YQ. 2013. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS One. 8:e57533.
- Richards AJ. 1973. The origin of Taraxacum agamospecies. Bot J Linn Soc. 66:189–211.
- Schütz K, Carle R, Schieber A. 2006. Taraxacum – a review on its phytochemical and pharmacological profile. J Ethnopharmacol. 107:313–323.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Timme RE, Kuehl JV, Boore JL, Jansen RK. 2007. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: identification of divergent regions and categorization of shared repeats. Am J Bot. 94:302–312.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.