Abstract
The complete mitochondrial genome (16,726 bp) of the winghead shark, Eusphyra blochii is presented. This species is exploited throughout parts of its range, and is currently listed as Near Threatened by the IUCN Red List. A phylogenetic analysis placed E. blochii within the Carcharhiniformes, as a sister taxon to Sphyrna lewini and S. zygaena.
Eusphyra is a monotypic genus of hammerhead shark in the family Sphyrnidae, order Carcharhiniformes. It is distinctive in having the most proportionally extreme narial separation among hammerhead sharks, which is suggested to increase odour resolution (Kajiura et al. Citation2005). Eusphyra blochii is found in shallow waters over continental and insular shelves of the Indian and Pacific Oceans, from the Persian Gulf to the Philippines, and from China to Australia (Simpfendorfer Citation2003; Ebert et al. Citation2013). Members of the family Sphyrnidae have been exploited by multiple fisheries leading to two species being listed as Endangered, two as Vulnerable and two as Near Threatened by the IUCN Red List (Pérez-Jiménez Citation2014). Life history traits coupled with fishing pressures make these species particularly vulnerable to over-exploitation. Eusphyra blochii is currently listed as Near Threatened due to heavy exploitation by fisheries in parts of its range (i.e. Gulf of Thailand, India and Indonesia) and light exploitation in Australian waters (Simpfendorfer Citation2003).
In this study, we determined the complete mitochondrial genome of E. blochii using a female specimen collected from Fog Bay, in the Timor Sea of northern Australia. A tissue sample from the specimen is deposited in the collection of Janine Caira at the University of Connecticut under field number AU-70 and extracted DNA is kept at the Hollings Marine Laboratory in Charleston, SC, under collection number GN5092. Total gDNA was extracted from liver tissue using the E.Z.N.A Tissue DNA Kit (Omega Bio-Tek, Inc., Norcross, GA) following the protocol of the manufacturer and the sequence was annotated using MitoAnnotator (Iwasaki et al. Citation2013). PCR primers and protocols are available upon request.
Twenty-eight shark and four batoid whole mitochondrial genome sequences were downloaded from GenBank and protein-coding regions of the 32 mitochondrial genomes were concatenated and aligned with corresponding regions from E. blochii. The complementary strand sequences were used for ND6, and incomplete stop codons of genes were removed from alignment. The final length of the protein-coding portion of the alignment was 11,424 bp. The dataset was subjected to maximum likelihood analysis using PAUP*4.0a145 (Swofford Citation2002) under the general-time reversible (GTR)+invariable sites (I)+gamma model with parameter values estimated from an initial parsimony analysis.
The complete mitochondrial genome (16,726 bp in length) of E. blochii has typical vertebrate mitochondrial gene arrangement (Anderson et al. Citation1981) consisting of 13 protein-coding genes, 22 tRNA genes, two rRNA genes and one control region (GenBank accession no. KJ128290). All genes are encoded on the H-strand, except for ND6 and eight tRNA genes, which are encoded on the L-strand.
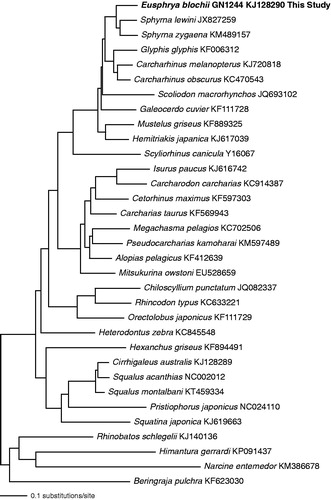
The maximum likelihood tree is given in . Based on this analysis and taxon sampling, E. blochii is placed within the Carcharhiniformes as sister taxon to S. lewini and S. zygaena, forming a monophyletic Sphyrnidae.
Funding information
This work was supported by National Science Foundation (DEB-1132229).
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperron IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290:457–465.
- Ebert DA, Fowler S, Compagno L. 2013. Sharks of the world: a fully illustrated guide. Plymouth (UK): Wild Nature Press.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Kajiura S, Forni J, Summers A. 2005. Olfactory morphology of carcharhinid and sphyrnid sharks: does the cephalofoil confer a sensory advantage? J Morphol. 264:25–263.
- Pérez-Jiménez JC. 2014. Historical records reveal potential extirpation of four hammerhead sharks (Sphyrna spp.) in Mexican Pacific waters. Rev Fish Biol Fish 24.2:671–683.
- Simpfendorfer CA. 2003. Eusphyra blochii. The IUCN Red List of Threatened Species. Version 2015.4; [cited 2016 Feb 2]. Available at: www.iucnredlist.org.
- Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland (MA): Sinauer Associates.