Abstract
Solanum commersonii Dunal is a well-known wild potato belonging to Solanaceae family and commonly used as materials for somatic hybridization due to various biotic and abiotic stress resistances. The complete chloroplast genome of S. commersonii was constituted by de novo assembly using a small amount of whole genome sequencing data. The chloroplast genome of S. commersonii was 155 525 bp in length, consisted of 86 013 bp of large single copy, 18 366 bp of small single copy region and 25 573 bp of a pair of inverted repeats. A total of 113 genes were annotated including 79 protein-coding genes, 30 tRNA genes and four rRNA genes. Maximum likelihood phylogenetic analysis with 14 Solanaceae species revealed that S. commersonii is much closely related to Solanum tuberosum and S. bulbocastanum.
Solanum commersonii is a wild diploid potato species, which is native in Uruguay, Argentina and Brazil. It has several desirable characteristics such as disease resistance to bacterial wilt and common scab, and abiotic stress resistance such as cold and frost tolerance (Carputo et al. Citation2009). Therefore, potato breeders have extensively used S. commersonii to make a somatic hybridization and bridge cross in order to introgress useful genetic traits from S. commersonii into a cultivated potato (Carputo et al. Citation1997, Citation2009, Citation2013). Somatic hybridization via protoplast fusion provides opportunity to overcome sexual barriers for interspecific gene transfer in potato breeding program (Kim-Lee et al. Citation2005; Yu, et al. Citation2013; Zhao et al. Citation2013). Aversano et al. (Citation2015) reported ∼830 Mb of draft genome sequences of S. commersonii, of which transposable elements comprise 44.5% of the genome. They also indicated that S. commersonii had diverged about ∼2.3 million years ago from domesticated potato and three duplication events for enrichment of gene families associated with biotic and abiotic stress resistance (Aversano et al. Citation2015).
Total genomic DNA was isolated from S. commersonii (PI320266) leaves provided by Highland Agriculture Research Institute (HARI), National Institute of Crop Science, Rural Development Administration, Pyeongchang, South Korea. Solanum commersonii line (PI320266) was originally introduced from Wisconsin Univ. and maintained in HARI as an in vitro plant. An Illumina paired-end (PE) genomic library was constructed according to the PE standard protocol (Illumina, San Diego, CA) and sequenced using an Illumina HiSeq2000 at Macrogen (http://www.macrogen.com/kor/), South Korea. Low-quality bases (Phred score less than 20) were trimmed, and then 3 Gbp of high-quality PE reads were assembled by a CLC genome assembler (ver. 4.06 beta, CLC Inc, Aarhus, Denmark) as described in Kim et al. (Citation2015). The representative chloroplast contigs were extracted and merged into a single draft sequence by comparison with the Solanum tuberosum (DQ386163.2) as a reference. The draft chloroplast sequence was further corrected by mapping raw PE reads. The chloroplast genes were predicted using the program DOGMA (Wyman et al. Citation2004) and BLAST searches.
The complete chloroplast genome of S. commersonii (GenBank accession no. KM489054) was 155 525 bp long, and the structure and gene features were typically identical to those of reported higher plants. It consisted of four well-defined regions such as 86,013 bp of large single copy (LSC), 18 366 bp of small single copy region (SSC) and 25 573 bp of a pair of inverted repeats (IRa and IRb). A total of 113 genes were annotated including 79 protein-coding genes, 30 tRNA genes and four rRNA genes. An overall GC content was 37.88%.
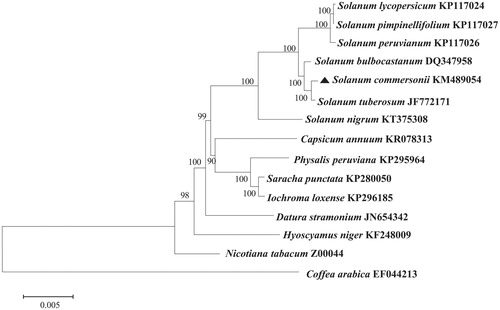
Phylogenetic relationship was analyzed using chloroplast coding sequences of S. commersonii and 14 published species in Solanaceae by a maximum likelihood method in MEGA 6.0 (Tamura et al. Citation2013). According to the phylogenetic tree, S. commersonii was much closely related to Solanum tuberosum and S. bulbocastanum, and separated with three tomato species among 14 species in Solanaceae, as expected ().
Disclosure statement
This work was carried out with the support of ‘Cooperative Research Program for Agriculture Science & Technology Development (Project title: Establishment of management system for improvement potato breeding program using potato germplasm, Project no. PJ00857304)’ Rural Development Administration, Republic of Korea.
References
- Aversano R, Contaldi F, Ercolano MR, Grosso V, Iorizzo M, Tatino F, Xumerle L, Dal Molin A, Avanzato C, Ferrarini A, et al. 2015. The Solanum commersonii genome sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives. Plant Cell. 27:954–968.
- Carputo D, Aversano R, Barone A, Di Matteo A, Iorizzo M, Sigillo L, et al. 2009. Resistance to Ralstonia solanacearum of Sexual Hybrids Between Solanum commersonii and S. tuberosum. Am J Potato Res. 86:196–202.
- Carputo D, Alioto D, Aversano R, Garramone R, Miraglia V, Villano C, Frusciante L. 2013) Genetic diversity among potato species as revealed by phenotypic resistances and SSR markers. Plant Genet. Resour C. 11:131–139.
- Carputo D, Barone A, Cardi T, Sebastiano A, Frusciante L, Peloquin SJ. 1997. Endosperm balance number manipulation for direct in vivo germplasm introgression to potato from a sexually isolated relative (Solanum commersonii Dun.). Proc Natl Acad Sci USA. 94:12013–12017.
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ. 2015. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 10:e0117159.
- Kim-Lee HY, Moon JS, Hong YJ, Kim MS, Cho HM. 2005. Bacterial wilt resistance in the progenies of the fusion hybrids between haploid of potato and Solanum commersonii. Am J Potato Res. 82:129–137.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yu Y, Ye WX, He L, Cai XK, Liu T, Liu J. 2013. Introgression of bacterial wilt resistance from eggplant to potato via protoplast fusion and genome components of the hybrids. Plant Cell Rep. 32:1687–1701.
- Zhao QX, Zhao BX, Zhang QQ, Yu B, Cheng LX, Jin R, Wang YP, Zhang JL, Wang D, Zhang F. 2013. Screening for chip-processing potato line from introgression of wild species' germplasms with post-harvest storage and chip qualities. Am J Potato Res. 90:425–439.