Abstract
The mitochondrial genome of the coral Porites lobata was sequenced using ezRAD. The assembled genome consists of 18,647 bp, including 13 protein-coding genes, two ribosomal RNA genes and two transfer RNA genes. The gene arrangement was consistent with other scleractinian coral mitochondrial genomes. The sequence was strikingly similar to Porites okinawensis, indicating the necessity for further systematic work to resolve phylogenetic relationships in the genus Porites.
Porites lobata (Dana Citation1846) is one of the most well-known, ecologically important reef building coral species in the world (Veron Citation2013); its geographic distribution extends from the Red Sea to the Eastern Pacific (Veron Citation2000), and colonies can live up to 1000 years (Brown et al. Citation2009), contributing substantially to the formation and maintenance of coral reefs (Baums et al. Citation2012). In Hawaii, P. lobata represents one of the most dominant coral species (Franklin et al. Citation2013). Although well studied, the taxonomy of Porites is highly contentious owing to phenotypic variation, plasticity and cryptic species as revealed by genetic and morphometric studies (e.g. Forsman et al. Citation2009; Prada et al. Citation2014; Forsman et al. Citation2015).
The mitochondrial genomes of animals share great similarity (e.g. Boore Citation1999), and highly conserved regions, such as the cytochrome c oxidase subunit I gene (COI), have been useful for a wide range of conservation, ecological, evolutionary and systematic studies (e.g. Hebert et al. Citation2003). The mitochondrial genome of scleractinian corals, however, is known to evolve extremely slowly (Shearer et al. Citation2002), and in many genera, including Porites, short mitochondrial markers such as COI have not been useful for closely related species. Therefore, sequencing the complete mitochondrial genome of P. lobata will enhance our understanding of the evolutionary relationships within the genus Porites.
Here we present the complete mitochondrial genome of P. lobata (GenBank access no. KU572435), assembled using next-generation sequencing. Small fragments of P. lobata samples were collected from Oahu, Hawaii (China Walls, Maunalua Bay: 21.2611°N, 157.7115°W, Site N, Maunalua Bay: 21.2765–21.2782°N, 157.7112–157.7116°W, Kewalo Basin: 21.9606°N, 157.8611°W and Lanikai: 21.3931°N 157.7149 W). DNA libraries were constructed using the Illumina TruSeq® Nano DNA kit, following the ezRAD Protocol modified from Toonen et al. (Citation2013). Individually barcoded samples were pooled, quality-checked and sequenced on an Illumina MiSeq® Analyzer at the Evolutionary Genetics Core Facility (Hawaii Institute of Marine Biology [HIMB], Kaneohe, HI). Quality-filtered reads were assembled to the mitochondrial genome of Porites okinawensis (GenBank access no. NC015644) using Geneious® v.6.0.5 (Biomatters Ltd. Auckland, New Zealand), as well as BWA v.0.7.12 (Li & Durbin Citation2009) to ensure the assembly quality and base calls. A consensus sequence was called using 0% majority option for coverage greater than 3 × (average 126×). Consensus sequences for 11 individuals were also called separately, which assembled 82.5–99.1% of the genome. Gene annotation was done using DOGMA (Wyman et al. Citation2004) and MITOS (Bernt et al. Citation2013), with additional verification of transfer RNA (tRNA) by tRNAscan-SE (Schattner et al. Citation2005) and RFam (Nawrocki et al. Citation2015). The leftover specimens are stored at Kewalo Marine Laboratory (sample ID: C6, C16, K2, M2, M7, M12, N1 and N3) and at HIMB (sample ID: PLob1, PLob2 and PLob3).
The length of P. lobata mitochondrial genome was 18,647 bp, the same length as that of P. okinawensis, with the base composition of A (22.2%), T (41.1%), C (12.9%) and G (23.7%), consistent with other scleractinian mitochondrial genomes that are A + T rich (Del Río-Portilla et al. Citation2016). The genome includes 13 protein-coding genes, two ribosomal RNA genes and two tRNA genes (tRNA-M and tRNA-W). The gene arrangement follows the same order as those of other Porites and scleractinian coral species (Lin et al. Citation2011; Del Río-Portilla et al. Citation2016). The pairwise sequence identity of P. lobata mitochondrial genome to P. okinawensis was 99.9%, well within the sequence variability of the mitochondrial genome observed among the 11 P. lobata individuals; approximately 99.8% (only 35 out of 18,647 of the sites were polymorphic). This highlights the need for further systematic work to determine species boundaries and geographic distributions of this recalcitrant group (Forsman et al. Citation2009).
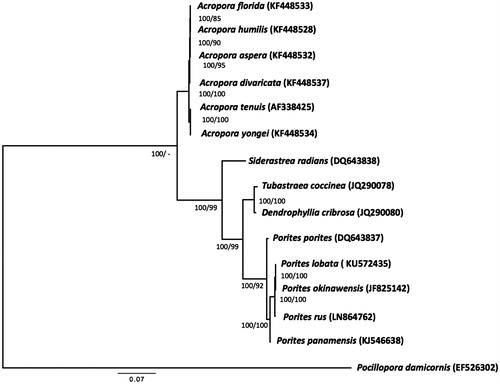
The phylogenetic tree () was constructed by the Bayesian and the maximum-likelihood methods using complete mitochondrial genomes of 15 scleractinian species. The tree supports clear phylogenetic relationships at the genus level. This mitochondrial genome (P. lobata) represents the fifth mitochondrial genome to be published for Porites, and provides additional insight into evolutionary relationships within the genus.
Figure 1. Phylogenetic tree of complete mitochondrial genomes from Porites lobata and other selected scleractinian coral species. The GenBank accession numbers are listed next to the species’ names. Numbers by each node represent the Bayesian posterior probability values (left) with 1.1 million generations obtained by MrBayes (Ronquist et al. Citation2012) and the maximum-likelihood bootstrap values (right) with 1000 replicates, obtained by PhyML (Guindon et al. Citation2010). Pocillopora damicornis was used as an outgroup for tree rooting.
Funding information
The authors wish to thank the Pauley Foundation for supporting the 2013 HIMB Pauley Program, which made this work possible. This work was also supported by Botany Department as part of a graduate class at the University of Hawaii at Manoa.
Acknowledgements
The authors thank for the support provided by Dr. Anthony S. Amend and Dr. Robert H. Richmond. The authors also appreciate Illumina Inc. for providing the reagents and supplies for sequencing. Coral samples were collected under Department of Land and Natural Resources permit SAP 2013-26 and 2015-06.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Baums IB, Boulay JN, Polato NR, Hellberg ME. 2012. No gene flow across the Eastern Pacific Barrier in the reef-building coral Porites lobata. Mol Ecol. 21:5418–5433.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler P. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phyl Evol. 69:313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Brown DP, Basch L, Barshis D, Forsman Z, Fenner D, Goldberg J. 2009. American Samoa’s island of giants: massive Porites colonies at Ta’u island. Coral Reefs. 28:735–735.
- Dana JD. 1846. United States exploring expedition during the years 1838–1842. Zoophytes. 7:1–740
- Del Río-Portilla MA, Vargas-Peralta CE, Paz-García DA, Lafarga De La Cruz F, Balart EF, García-de-León FJ. 2016. The complete mitochondrial DNA of endemic Eastern Pacific coral (Porites panamensis). Mitochondrial DNA. 27:738–739.
- Forsman Z, Wellington GM, Fox GE, Toonen RJ. 2015. Clues to unraveling the coral species problem: distinguishing species from geographic variation in Porites across the Pacific with molecular markers and microskeletal traits. Peer J. 3:e751.
- Forsman ZH, Barshis DJ, Hunter CL, Toonen RJ. 2009. Shape-shifting corals: molecular markers show morphology is evolutionarily plastic in Porites. BMC Evol Biol. 9:45.
- Franklin EC, Jokiel PL, Donahue MJ. 2013. Predictive modeling of coral distribution and abundance in the Hawaiian Islands. Mar Ecol Prog Ser. 481:121–132.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Hebert PDN, Cywinska A, Ball SL, de Waard JR. 2003. Biological identifications through DNA barcodes. Proc R Soc B: Biol Sci. 270:313–321.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25:1754–1760.
- Lin MF, Luzon KS, Licuanan WY, Ablan-Lagman MC, Chen CA. 2011. Seventy-four universal primers for characterizing the complete mitochondrial genomes of scleractinian corals (Cnidaria; Anthozoa). Zool Stud. 50:513–524.
- Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, Floden EW, Gardner PP, Jones TA, Tate J, et al. 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43:D130–D137.
- Prada C, DeBiasse MB, Neigel JE, Yednock B, Stake JL, Forsman ZH, Baums IB, Hellberg ME. 2014. Genetic species delineation among branching Caribbean Porites corals. Coral Reefs. 33:1019–1030.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server):W686–W689.
- Shearer TL, van Oppen MJH, Romano SL, Wörheide G. 2002. Slow mitochondrial DNA sequence evolution in the Anthozoa (Cnidaria). Mol Ecol. 11:2475–2487.
- Toonen RJ, Puritz JB, Forsman ZH, Whitney JL, Fernandez-Silva I, Andrews KR, Bird CE. 2013. ezRAD: a simplified method for genomic genotyping in non-model organisms. PeerJ. 1:e203.
- Veron J. 2013. Overview of the taxonomy of zooxanthellate Scleractinia. Zool J Linn Soc. 169:485–508.
- Veron JEN. 2000. Corals of the world. Australia: Australian Institute of Marine Science and CRR Qld Pty Ltd.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
