Abstract
Ostericum koreanum Kitagawa is an important herbal medicine, whose taxonomic status has been changed to Angelica reflexa as a new species. This study generated the complete chloroplast genome sequence of O. koreanum, and reconsidered its molecular taxonomic status in Angelica by comparing it with related species. The length of the complete chloroplast genome was 147,282 bp, and there were four structures that included the large single copy region (93,185 bp), the small single copy region (17,663 bp) and the duplicated inverted regions (18,217 bp of each). Based on its phylogenetic trees, O. koreanum was grouped by high bootstrap value with the Angelica species. This result proved that O. koreanum is included in Angelica. Therefore, this chloroplast genome data generated for the first time a valuable genetic resource for the discrimination of herbal materials, phylogeny, and evolution.
Ostericum koreanum Kitagawa, has been used as a beneficial herbal medicinal plant with rejuvenating effects, belongs to the family Apiaceae (Kim et al. Citation2014). This species has been known as a controversial taxon, whose classification has been changed several times. Maximowicz (Citation1886) reported a new species as Angelica koreana, and this species was then transferred to the genus Ostericum and O. grosseserrata by Kitagawa (Citation1936, Citation1971), respectively. Recently, however, Lee et al. (Citation2013) reported that O. koreanum was reclassified to Angelica reflexa as a new species. In this study, we determined the complete chloroplast genome of O. koreanum for the first time, and estimated its molecular taxonomic status in Angelica.
The plant sample of O. koreanum was collected from the herbal garden of the National Center for Herbal Medicine Resources in Korea. The genomic DNA was extracted from dried leaf and assembled to Illumina paired-end library. Genomic sequencing was performed using Illumina Hiseq 2000 instrument (Illumina, San Diego, CA), and assembled by CLC genomic assembler (v. beta 4.6, CLC Inc., Rarhus, Denmark). The gene annotation was conducted to DOGMA (http://dogma.ccbb.utexas.edu/) and manual curation using BLAST. The voucher specimen was preserved in the National Center for Herbal Medicine Resources.
The total size of the assembled chloroplast genome of O. koreanum was 147,282 bp (accession no. KT852844), and the genome reads were 215× of average coverage. The genome structure consisted of the typical four parts, which were large single copy region (LSC) of 93,185 bp, small single copy region (SSC) of 17,663 bp and a pair of inverted regions (IR) of 18,217 bp each. The GC content was 37.54%. The genome had a total of 113 genes, including 80 protein-coding genes among which nine (ndhB, orf42, orf56, rps7, rps12, ycf1, ycf2, ycf15 and ycf68) were duplicated in the IR region. There were 29 tRNA genes distributed throughout the genome, with 22 in the LSC, one in the SSC and six in the individual IRs. There were four rRNA, the rrn4.5, rrn5, rrn16 and rrn23 genes, distributed in only the IR regions.
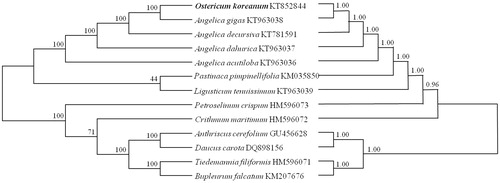
To determine the taxonomic status of O. koreanum within Angelica, phylogenetic analyses based on maximum parsimony (MP) and maximum likelihood (ML) were performed from the genome sequences of 11 taxa, consisting of genera Angelica, Ligusticum, Glehnia, Peucedanum and Saposhnikovia. These sequences were aligned using MAFFT (http://mafft.cbrc.jp/alignment/software/). The MP tree was constructed by 10 random additions, tree-bisection-reconnection (TBR), and 1000 bootstrap replications in MEGA6 (Tamura et al. Citation2013). The ML tree was produced by GARLI Web Services (www.molecularevolution.org), with general time reversible parameters based on gamma distribution and 1000 replicates.
Both the MP and the ML trees were similar, as indicated by their bootstrap support values on the branches and topology of trees (). In MP tree, O. koreanum showed monophyletic relation with Angelica species, supported by high statistical values of nearly 100%, and in ML tree O. koreanum showed a close relationship with the Angelica species. Therefore, our data of complete genomic sequences proved the study by Lee et al. (Citation2013) that O. koreanum is included in genus Angelica. This chloroplast genome data could be supported to discriminate between genuine species for herbal medicine and adulterations, to analyze the relation of closely related taxa, and to study phylogeny and evolution.
Funding information
This research was supported by a grant (13171MFDS419) from the Ministry of Food and Drug Safety of the Republic of Korea in 2014.
Disclosure statement
The authors report that they have no conflicts of interest.
References
- Kim CM, Lee YJ, Kim IR, Shin JH, Kim YI. 2014. Coloured illustrations for discrimination of herbal medicine. Seoul, Republic of Korea: Academy Book.
- Kitagawa M. 1936. Ostericum and Angelica from Manchuria and Korea. J Jpn Bot. 12:229–246.
- Kitagawa M. 1971. On the syntype specimens of Angelica koreana Maximowicz. J Jpn Bot. 46:367–372.
- Lee BY, Kwak MH, Han JE, Jung EH, Nam GH. 2013. Ganghwal is a new species, Angelica reflexa. J Species Res. 2:245–248.
- Maximowicz CJ. 1886. Diagnoses plantarum novarum Asiaticarum. Bull Acad impérial sci St.-Pétersbourg 31:48–54.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.