Abstract
The sword prawn Parapenaeopsis hardwickii (Miers) (Crustacea, Decapoda, Penaeidae) is an important commercial fishery species, distributed in the East China Sea. In this study, we described the complete mitochondrial genome of P. hardwickii. The genome is 15,922 bp in length, encoding the standard set of 13 protein-coding genes, 22 tRNA genes and two rRNA genes, with circular organization. The overall A + T content is 67.20%; nucleotide frequency of the gene is as follows: A, 35.01%; C, 20.83%; G, 11.98%; T, 32.19%. The gene order of P. hardwickii is the same as Penaeus monodon and Fenneropenaeus chinensis, and it mainly retains as the Penaeidae ground pattern. The complete mitogenome sequence information of P. hardwickii would play an important part for further studies on molecular systematics and population genetics.
The sword prawn Parapenaeopsis hardwickii (Crustacea: Decapoda: Penaeidae: Parapenaeus) inhabits sandy ocean bottoms at depths of 5–90 m in the coastal seawater of the Indo-West Pacific Ocean from Pakistan to East China Sea (Tzeng Citation2007). The life history of the sword prawn, with an offshore planktonic larval phase, estuarine post-larval and juvenile phases, and offshore adult and spawning phases (Dall et al. Citation1990), makes it be a very abundant and highly valued species in the Xiangshan Harbour aera (East China Sea) and waters adjacent to Taiwan (Wu Citation1982; Song & Ding Citation1993). Although it is a valued and high-yield marine resource, the resources of marine prawns, including P. hardwickii, decreased clearly in the East China Sea and Taiwan due to overcatching and the breed descending in past decades (Tzeng Citation2004). Because of its economic and ecological importance, and less information for population genetics, we need more molecular data and genome study for this species.
The P. hardwickii sample was collected from Xiangshan Harbour aera, Zhejiang Province, China. The total genomic DNA was extracted from abdominal muscle tissue of the specimen stored in 70% ethanol, which is stored in Key Laboratory of Sustainable Development of Marine Fisheries, Ministry of Agriculture, PR China, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences. The complete mitogenome of P. hardwickii is 15,922 bp in length (GenBank accession no. KU302814) and contains 13 protein-coding genes (nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad5, nad4, nad4L, nad6, cob and nad1), two ribosomal RNA (rRNA) genes (12S rRNA and 16S rRNA) and 25 transfer RNA genes. Eight protein-coding genes are started with the ATG typical crustacean codons and the nad2, atp8 and nad6 genes shared ATT codons, while cox1 and nad4 have the ACG and GTG condons, respectively. Seven genes terminated open reading frames with a complete stop condon, either TAA (nad2, cox1, atp8, atp6, nad4 and nad4L) or TAG (cob), while the remaining genes have incomplete stop condon T. The overall GC content is 32.81%, which is within normal limits of crustacean mitochondrial DNAs. Nucletide frequency of the gene is as follows: A, 35.01%; C, 20.83%; G, 11.98%; T, 32.19%. All the 25 typical tRNAs possess a complete clover leaf secondary structure, ranging from 63 bp to 106 bp. These results were obtained by RNA scan-SE 1.21 (Lowe & Eddy Citation1997) and DOGMA (Wyman et al. Citation2004). There was no completely identical block compared to those of P. monodon (NC_002184) and F. chinensis (NC_009679).
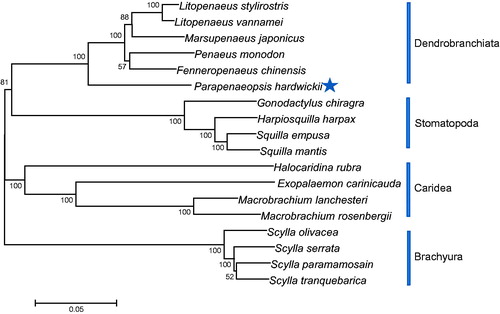
The phylogenetic position of P. hardwickii within Dendrobranchiata was studied with the concatenated amino acid sequences of 13 PCGs using maximum likelihood (ML) methods. The results showed that P. hardwickii firstly grouped with species of Penaeidae, and closely related to M. japonicas and F. chinensis. Phylogenetic analyses consistent with the classification results depended on the morphological characters (). We have the confidence to construct phylogenetic trees, based on the complete the mitochondrial genomes.
Figure 1. Phylogenetic tree for decapod relationships based on the dataset of 13 concatenated mitochondrial PCGs. Branch lengths and topologies came from the maximum-likelihood (ML) analyses. Numbers besides the nodes specify bootstrap percentages from ML. Four stomatopods served as outgroups. Sequence data for phylogenetic analyses used in the study were as follows: Scylla olivacea (NC_012569), Scylla tranquebarica (NC_012567), Scylla paramamosain (NC_012572), Scylla serrata (NC_012565), Exopalaemon carinicauda (NC_012566), Macrobrachium lanchesteri (NC_012217), Macrobrachium rosenbergii (NC_006880), Marsupenaeus japonicus (NC_007010), Fenneropenaeus chinensis (NC_009679), Litopenaeus stylirostris (NC_012060), Litopenaeus vannamei (NC_009626), Gonodactylus chiragra (NC_007442), Harpiosquilla harpax (NC_006916), Squilla empusa (NC_007444), and Squilla mantis (NC_006081), Penaeus monodon (NC_002184), Halocaridina rubra (NC_008413).
Funding information
This work is supported by a project “Aquatic Germplasm Resources Platform Operation Service” for National Science and Technology Basic Conditions Platform, National Shrimp Industry Technology System (CARS-47).
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Dall W, Hill BJ, Rothlisberg PC, Sharples DJ. 1990. The biology of the Penaeidae. London: Academic Press.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Song HT, TM Ding. 1993. A comparative study on fishery biology of main economic shrimps in the north of East China Sea. J Zhejiang Coll Fish. 12:240–248.
- Tzeng TD. 2004. Stock identification of sword prawn Parapenaeopsis hardwickii in the East China Sea and Taiwan Strait inferred by morphometric variation. Fisheries Sci. 70:758–764.
- Tzeng TD. 2007. Population structure of the sword prawn (Parapenaeopsis hardwickii) (Decapoda: Penaeidae) in the East China Sea and waters adjacent to Taiwan inferred from the mitochondrial control region. Zool Stud. 46:561–568.
- Wu B. 1982. Some problems on circulation study in Taiwan Strait. Taiwan Strait 1:1–7.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
