Abstract
The complete chloroplast genome sequence of Angelica gigas, a traditional herbal plant used in treating diseases, was obtained by de novo assembly using illumina sequencing data (Illumina Inc., San Diego, CA). The circular molecule of the genome was constructed of four parts, with a size of 146,916 bp in total – a large single copy (LSC) region of 93,118 bp, a small single copy (SSC) region of 17,582 bp and two inverted repeat (IRa and IRb) regions of 18,108 bp each. There were a total of 113 annotated genes, including 80 protein-coding genes, 29 tRNA genes and four rRNA genes. The phylogenetic result acquired through maximum parsimony analysis showed that A. gigas is closely related with A. decursiva and Seseli montanum.
Angelica gigas Nakai belongs to the family Apiaceae, and is a perennial herb found in Korea, China and Japan (Zehui & Watson Citation2005). The roots of this plant, called Dang-gui in Korea (Korea Food and Drug Administration, Citation2013), have been well used as a traditional medicinal plant for gynaecological diseases, haematopoiesis and anti-inflammation (Ahn et al. Citation1996; Yoon et al. Citation2007). The material plants for Dang-gui differ among countries depending on their regulations, with A. sinensis (Oliv.) Diels in China (Pharmacopoeia Commission of the People’s Republic of China, Citation2010) and A. acutiloba Kitagawa or A. acutiloba Kitagawa var. sugiyamana Hikino in Japan (The Ministry of Health, Labour and Welfare, Citation2011). However, A. acutiloba and A. acutiloba var. sugiyamana are prescribed as Il-dang-gui in Korea. Because of the similar medicinal names and closely related species in the genus Angelica among herbal materials, mixes or misuses could occur in the market place. We, therefore, assembled the complete chloroplast genome of A. gigas, to provide a genomic resource for fair trade and harmony in the regulation of herbal medicines through correct discrimination among species.
The leaves of A. gigas was obtained from a farmhouse in Jecheon-si, Korea (37°11'22.10''N, 128°15'32.99''E), and extracted by DNeasy Plant Mini Kit (QIAGEN Inc., Valencia, CA). The isolated genomic DNA was manufactured to 300 bp paired-end (PE) library using the Illumina Hiseq library kit (Illumina, San Diego, CA), and sequenced by Illumina genome analyzer (Hiseq2000). Acquired high-quality reads were conducted by de novo assembly using CLC genome assembler (ver. 4.06 beta, CLC Inc, Rarhus, Denmark). The assembled structures and genes of the complete chloroplast genome were annotated by DOGMA (http://dogma.ccbb.utexas.edu/) (Wyman et al. Citation2004), and manually corrected by Blast search. The chloroplast genome map was drawn with OGDRAW v 1.2 (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2007). The complete chloroplast genome sequence was deposited in GenBank under accession no. KT963098. Sample was deposited at the National Center for Herbal Medicine Resources, NIFDS, and MFDS, of Korea under accession no. 13E-39.
The chloroplast genome of A. gigas was a circular molecule with a size of 146,916 bp comprising four structures, with 93,118 bp in the large single copy (LSC) region, 17,582 bp in the small single copy (SSC) region and 18,108 bp in each of duplicated inverted repeat (IRa and IRb) regions. The overall GC content was 37.56%, with the GC contests of the IRs the highest at 44.81% and followed by those of the LSC (35.97) and the SSC (31.01). The chloroplast genome contained a total of 113 genes – 80 protein-coding genes, 29 tRNA genes and four rRNA genes.
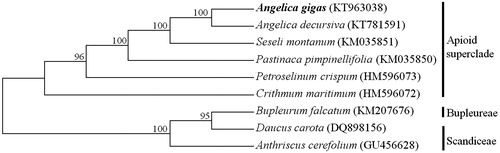
To analyze the phylogenetic relationships between A. gigas and related species in Apiaceae, the chloroplast genome sequences of a total of nine taxa were aligned by MAFFT (http://mafft.cbrc.jp/alignment/software/) (Katoh et al. Citation2002). Maximum parsimony (MP) analysis was performed based on tree bisection reconnection (TBR) branch swapping with 1000 replications in MEGA6 (Tamura et al. Citation2013). The phylogenetic tree was divided into two groups, which were Apioid superclade with Angelica, and two other tribes, Bupleureae and Scandiceae (). A. gigas showed a close relationship to the same genus species A. decursiva and Seseli montanum.
Funding statement
This research was supported by a grant (13171MFDS419) from the Ministry of Food and Drug Safety of the Republic of Korea in 2014.
Disclosure statement
The authors declare that there is no conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- Ahn KS, Sim, WS, Kim, HM, Han, SB, Kim, IH. 1996. Immunostimulating components from the root of Angelica gigas Nakai. Kor J Pharmacogn. 27:254–261.
- Korea Food and Drug Administration. 2013. The 10th Korean Pharmacopoeia (KP). Seoul, Korea Food and Drug Administration, 26 Korean Pharmacopoeia.
- Katoh M, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Lohse M, Drechsel O, Bock R. 2007. Organellar Genome DRAW (OGDRAW): a tool for easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Gnet. 52:267–274.
- Pharmacopoeia Commission of the People's Republic of China. 2010. Pharmacopoeia of the People's Republic of China. Beijing: China Medical Science and Technology Press.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- The Ministry of Health, Labour and Welfare. 2011. The Japanese Pharmacopoeia Sixteenth Edition. Japan: Labour and Welfare Ministerial Notification No. 65.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yoon TS, Cheon MS, Lee DY, Moon BC, Lee HW, Choo BK, Kim HK. 2007. Effects of root extracts from Angelica gigas and Angelica acutiloba on inflammatory mediators in mouse macrophages. J Appl Biol Chem. 50:264–269.
- Zehui P, Watson MF. 2005. Angelica. Flora of China, vol. 14: 165. St. Louis (MO), Cambridge (MA): Missouri Botanical Garden, Harvard University Herbaria; [cited 2015 Oct 27]. Available from: http://www.efloras.org/florataxon.aspx?flora_id=2&taxon_id=200015368.