Abstract
The complete mitochondrial genome was sequenced in two individuals of stone char Salvelinus kuznetzovi. The genome sequences are 16,654 bp in size, and the gene arrangement, composition and size are very similar to the salmonid fish genomes published previously. The low level of sequence divergence detected between the genome of S. kuznetzovi and the GenBank complete mitochondrial genomes of the white char S. albus (KT266870 and KT266871), the Northern Dolly Varden char S. malma (KJ746618) and the Arctic char S. alpinus (AF154851) may likely be due to recent divergence and/or historical hybridization and interspecific replacement of mtDNA.
The stone char Salvelinus kuznetzovi (Taranetz Citation1933) is an endemic of the Kamchatka River with uncertain taxonomic status. Savvaitova and Maksimov (Citation1970) included S. kuznetzovi in the S. alpinus complex. Glubokovsky (Citation1995) based on morphological data suggested that S. kuznetzivi represents a resident form of the white char S. albus. However, Sheiko and Fedorov (Citation2000) as well as Bogutskaya and Naseka (Citation2004) considered the stone char as a separate species. No genetic data are available to confirm the species identity of S. kuznetzovi.
We have sequenced two complete mitochondrial (mt) genomes of S. kuznetzovi (GenBank accession nos. KU674351 and KU674352) from the Azabachye lake creek, Kamchatka, Russia (56°08′30″N, 161°48′00″E), using primers designed with the program mitoPrimer_V1 (Yang et al. Citation2011). The fish specimens are stored at the museum of the A. V. Zhirmunsky Institute of Marine Biology, Vladivostok, Russia (www.museumimb.ru) under accession nos. SK1 and SK2. The size of the genome is 16,654 bp and the gene arrangement, composition and size are very similar to the salmonid fish genomes published previously. There were four single nucleotides and no any length differences between the haplotypes SK1 and SK2; the total sequence divergence (Dxy) was 0.0002 ± 0.0001.
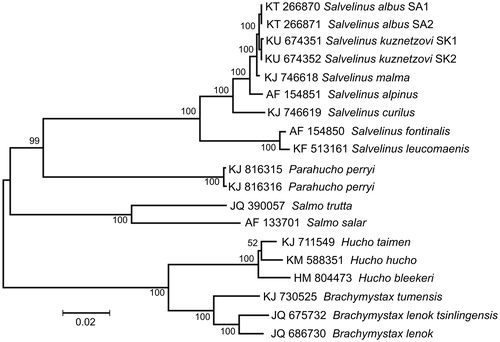
The comparison of mt genomes now obtained with other complete mt genomes available in GenBank for the family Salmonidae including genera Salvelinus, Parahucho, Salmo, Hucho and Brachymystax reveals a close affinity of S. kuznetzovi to other Salvelinus species () with a very low level of sequence divergence between our specimens (SK1 and SK2) and the complete mt genome of the white char S. albus (Dxy = 0.0010 ± 0.0003; Balakirev et al. Citation2015) and the Northern Dolly Varden char S. malma (Dxy = 0.0023 ± 0.0003; Balakirev et al. Citation2016). The divergence was higher (Dxy = 0.0079 ± 0.0006) between S. kuznetzovi and S. alpinus (Dxy = 0.0090 ± 0.0010; Doiron et al. Citation2002), but still too low for considering them as separate species. The low level of sequence divergence among S. kuznetzovi, S. albus, S. malma and S. alpinus could be explained by recent divergence and/or historical hybridization and interspecific replacement of mtDNA, as it has been found for other char species (e.g. Bernatchez et al. Citation1995).
Figure 1. Maximum likelihood tree for the stone char Salvelinus kuznetzovi specimens SK1 and SK2, and the GenBank representatives of the family Salmonidae. The tree is constructed using whole mitogenome sequences. The tree is based on the General Time Reversible + gamma + invariant sites (GTR + G + I) model of nucleotide substitution. The numbers at the nodes are bootstrap percent probability values based on 1000 replications.
Funding information
This work was supported by the Bren Professor Funds at the University of California, Irvine, USA (mitochondrial genome sequencing); and the Russian Science Foundation under Grant number 14-50-00034 (data analysis).
Disclosure statement
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors alone are responsible for the content and writing of the paper.
The authors declare no financial interest or benefit from the direct applications of this research. The authors report that they have no conflicts of interest.
References
- Balakirev ES, Parensky VA, Kovalev MY, Ayala FJ. 2015. Complete mitochondrial genome of the white char Salvelinus albus (Salmoniformes, Salmonidae). Mitochondrial DNA. 1–2. [Epub ahead of print]. doi: 10.3109/19401736.2015.1079890.
- Balakirev ES, Romanov NS, Ayala FJ. 2016. Complete mitochondrial genomes of the Northern (Salvelinus malma) and Southern (Salvelinus curilus) Dolly Varden chars (Salmoniformes, Salmonidae). Mitochondrial DNA. 27:1016–1017.
- Bernatchez L, Glémet H, Wilson CC, Danzmann R. 1995. Introgression and fixation of Arctic charr (Salvelinus alpinus) mitochondrial genome in an allopatric population of brook trout (Salvelinus fontinalis). Can J Fish Aquat Sci. 52:179–185.
- Bogutskaya NG, Naseka AM. 2004. Catalogue of agnathans and fishes of fresh and brackish waters of Russia with comments on nomenclature and taxonomy. Moscow: KMK Scientific Press Ltd.
- Doiron S, Bernatchez L, Blier PU. 2002. A comparative mitogenomic analysis of the potential adaptive value of Arctic charr mtDNA introgression in brook charr populations (Salvelinus fontinalis Mitchill). Mol Biol Evol. 19:1902–1909.
- Glubokovsky MK. 1995. Evolutionary biology of salmonid fishes. Moskow: Nauka.
- Savvaitova KA, Maksimov VA. 1970. Stone char from the Kamchatka river watershed. Biol Sci. 5:7–20.
- Sheiko BA, Fedorov VV. 2000. Class Cephalaspidomorphi – Lampreys. Class Chondrichthyes – Cartilaginous Fishes. Class Holocephali – Chimaeras. Class Osteichthyes – Bony Fishes. In: Moiseev RS, Tokranov AM, editors. Catalog of vertebrates of Kamchatka and Adjacent Waters. Petropavlovsk-Kamchatsky: Kamchatskiy Petchatniy Dvor. p. 7–69.
- Taranets AY. 1933. Some new freshwater fishes from the Far Eastern region. Dokl Akad Nauk SSSR. 2:83–85.
- Yang CH, Chang HW, Ho CH, Chou YC, Chuang LY. 2011. Conserved PCR primer set designing for closely-related species to complete mitochondrial genome sequencing using a sliding window-based PSO algorithm. PLoS One. 6:e17729.
