Abstract
We report the complete chloroplast DNA (cpDNA) of Arabidopsis lyrata (Brassicaceae), a less studied relative of A. thaliana, by employing next-generation sequencing reads and de novo assembly. The length of the closed circular cpDNA is 154,604 bp with a typical quadripartite structure. The genome is composed of one large single copy and one small single copy regions of 84,209 bp and 17,871 bp, respectively, and separated by a pair of inverted repeats of 26,262 bp in length. The overall GC content is 36.35% and the GC content of the LSC, IRs and SSC regions are 34.12%, 42.30% and 29.38%, separately. The gene content and the number for A. lyrata are the same as other published species in Brassicaceae with 112 annotated known unique genes including 78 protein-coding genes, 30 tRNA genes and four rRNA genes. The complete cpDNA of A. lyrata will provide valuable molecular resources for further phylogenetic and evolutionary analysis in the model Arabidopsis genus.
As a very close relative to model species Arabidopsis thaliana (Brassicaceae), the full nuclear genome of A. lyrata has been published (Hu et al. Citation2011). Despite their close relation, some very important biological differences exist between A. lyrata and A. thaliana. Most notably the mating system for A. thaliana is strict selfing, whereas A. lyrata is a perennial outcrossing diploid species (Ross-Ibarra et al. Citation2010). Furthermore, the complete nuclear genome size and chromosome number are vastly different between the two species with A. lyrata possessing eight chromosomes for a nuclear genome that is an about 1.5–2 times larger than A. thaliana with only five chromosomes. These differences are even more striking when considering that the divergence between these lineages is estimated at five million years ago (Hu et al. Citation2011). While comparative genomic studies have taken place between A. lyrata and A. thaliana the lack of a complete chloroplast genome for A. lyrata has limited the research that can be done using this effectively non-recombinant, uniparentally inherited genome. For instance, the plant cpDNA has been used in areas of research as diverse as molecular systematics (Jansen et al. Citation2007; Wang et al. Citation2010; Wu & Ge Citation2012), studying biogeographical relationships among populations (Wang et al. Citation2011), plant DNA barcoding (Group CPBOL et al. Citation2011) and plant genetic transformation (Cui et al. Citation2011). In this study, we report it’s the complete cpDNA of A. lyrata by employing the published nuclear genome data.
By downloading the reads from NCBI accession DRR013372 (Hu et al. Citation2011), its accession number is CS22696 that was deposited at the Arabidopsis Biological Resource Center; it was collected as a forced inbred strain named MN47 in Michigan, USA); the full chloroplast genome was assembled following the method used in Wu (Citation2015) in the CLC workbench (ver. 7.01 beta, CLC Inc, Aarhus, Denmark). This finished sequence was also validated by mapping the raw PE reads back to itself. The genome annotation and structural features of this genome were predicted using the method from Wu and Ge (Citation2016). The deposited NCBI accession number of A. lyrata is KU559924.
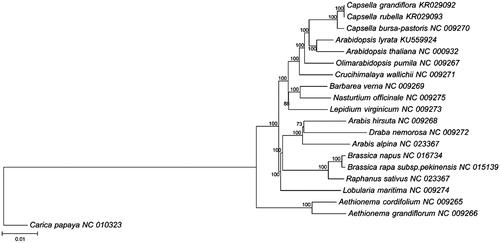
The complete cpDNA for A. lyrata has a total length of 154,604 bp with a characteristic quadripartite structure, consisting of an LSC region of 84,209 bp, two IR regions of 26,262 bp and an SSC region of 17,871 bp. This typical structure is conserved and identical to all other published cpDNA in all Brassicaceae species (Wu Citation2015). The chloroplast genome has a GC content of 36.35% and 112 coding genes, including 78 protein-coding genes, 30 tRNA genes and four rRNA genes. All four rRNA genes are located in the IR regions. Twenty-three tRNA genes are located in the two single copy regions, whereas the other seven are located in the IR regions. Eighteen genes contain introns: ycf3, rps12 and clpP contain two introns, and the rest of the genes contain a single intron. Six of those 18 intron containing genes are tRNA genes. Rps12 is trans-spliced, with one of its exons in the LSC region (5′ end) and the other two exons in the IR region (3′ end) separated by an intron. Phylogenetic analysis using the whole cpDNA alignment from 18 published Brassicaceae species including A. lyrata and one outgroup Carica papaya, was conducted using neighbour-joining (NJ) in MEGA 6.0 (Tamura et al. Citation2013). Our phylogenetic analysis confirms the relationship between A. lyrata and A. thaliana within the Brassicaceae ().
Figure 1. Molecular phylogeny of Arabidopsis lyrata and 18 species from Brassicaceae was based on complete cpDNA sequences. Sequence data was downloaded from GenBank database and the phylogenic tree was constructed by neighbor-joining method with 500 bootstrap replicates in MEGA 6 (Tamura et al. Citation2013). The GenBank accession number of each species used for tree construction is listed after the species name, Carica papaya (NC_010323) was used as the out-group species.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Cui C, Song F, TanY, Zhou X, Zhao W, Ma F, Liu Y, Hussain J, Wang Y, Yang G, et al. 2011. Stable chloroplast transformation of immature scutella and inflorescences in wheat (Triticum aestivum L.). Acta Biochim Biophys Sin. 43:284–291.
- Group CPBOL, Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, Chen ZD, Zhou SL, Chen SL, et al. 2011. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc Natl Acad Sci USA. 108:19641–19646.
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 43:476–481.
- Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 104:19369–19374.
- Ross-Ibarra J, Wright SI, Foxe JP, Kawabe A, DeRose-Wilson L, Gos G, Charlesworth D, Gaut BS. 2010. Patterns of polymorphism and demographic history in natural populations of Arabidopsis lyrata. PLoS One. 3:e2411.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang L, Qi XP, Xiang QP, Heinrichs J, Schneider H, Zhang XC. 2010. Phylogeny of the paleotropical fern genus Lepisorus (Polypodiaceae, Polypodiopsida) inferred from four chloroplast genome regions. Mol Phylogenet Evol. 54:211–225.
- Wang L, Wu ZQ, Bystriakova N, Ansell SW, Xiang QP, Heinrichs J, Schneider H. 2011. Phylogeography of the Sino-Himalayan fern Lepisorus clathratus on “the roof of the world”. PLoS One. 6:e25896.
- Wu ZQ. 2015. The complete chloroplast genome of Capsella rubella. Mitochondrial DNA Part A. [Epub ahead of print]. DOI: 10.3109/19401736.2015.1038804.
- Wu ZQ, Ge S. 2012. The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol Phylogenet Evol. 62:573–578.
- Wu ZQ, Ge S. 2016. The whole chloroplast genome of wild rice (Oryza australiensis). Mitochondrial DNA Part A. 27:1062–1063.
