Abstract
We describe the complete mitochondrial genome sequence of Oncorhynchus masou formosanus, which is a critically endangered landlocked salmon of Taiwan by using next-generation sequencing. The circle genome (16,653 bp) has the typical vertebrate mitochondrial gene arrangement, consisting of 13 protein-coding genes, 22 tRNA genes, two rRNA genes and a non-coding control region. The overall base composition of O. m. formosanus is 28.6% for A, 26.8% for T, 16.5% for G and 28.1% for C, with a slight AT bias of 55.4%.
The landlocked salmon of Taiwan, Oncorhynchus masou formosanus (Jordan & Oshima Citation1919), was listed as Critically Endangered by the IUCN in 1996 (Kottelat Citation1996), which only distributed in the upstream of Dajia River (Hsu et al. Citation2010). The total number of O. m. formosanus has increased from 200 to around 3000 through the efforts of repopulation and habitat recovery (Chung et al. Citation2008). Since the species status of O. m. formosanus is conflict (Numachi et al. Citation1990; Gwo et al. Citation2008) and the characteristics of landlocked and southernmost distribution of salmonids has been considered, mitochondrial genome may provide additional reference to not only conservation issue, but also taxonomy and phylogeography study.
Here, we determined the complete mitochondrial genome sequence with all genes annotated of O. m. formosanus that was deposited in GenBank with accession number KU523579. The two specimens of O. m. formosanus were used in this study, one was from wild environment and another was cultivated individual of Hatchery and Conservation Center of Formosa Salmon, Shei-Pa National Park. The number of collection licenses is 103020–103033. The mtDNAs were extracted and prepared according to manufacturer’s instructions of Illumina MiSeq platform (Illumina Inc., San Diego, CA). The paired-end sequencing reads (250 bases) of mitochondrial genome were assembled by CLC Genomics Workbench version 5.5.1 (CLC-bio, Aarhus, Denmark). The annotation of protein-coding genes and rRNA genes were determined by online software MitoAnnotator (Iwasaki et al. Citation2013). To elucidate the phylogenetic relationship between O. m. formosanus and other closely related species, 16 mitochondrial genomes were used for phylogenetic analysis. The complete mitochondrial genome sequences of 15 other species are downloaded from GenBank and the names and GenBank accession numbers of these species are as follows: O. mykiss (NC_001717), O. tshawytscha (NC_002980), O. clarkia henshawi (NC_006897), O. nerka (NC_008615), O. masou ishikawae (NC_008746), O. masou masou (NC_008747), O. masou Biwa (NC_009262), O. kisutch (NC_009263), Salmo trutta trutta (NC_010007), O. gorbuscha (NC_010959), O. keta (NC_017838), Brachymystax lenok (NC_018341), B. lenok tsinlingensis (NC_018342), S. trutta (NC_024032) and O. mykiss x S. salar (NC_026537).
The complete mitochondrial genomes of these two specimens were identical (16,653 bp in length) which consists of 13 protein-coding genes, 22 tRNA genes, two rRNA genes and one control region (CR). The arrangement of all genes was identical to that of other Cypriniformes (Yang et al. Citation2013). Most of the genes are encoded on the heavy strand (H-strand), except for the eight tRNA genes (tRNA-Gln, –Ala, –Asn, –Cys, –Tyr, –Ser, –Glu and –Pro) and one protein-coding gene (ND6). Its overall base composition is 28.6% for A, 26.8% for T, 16.5% for G and 28.1% for C, with relatively lower of GC and AT bias of 55.4%.
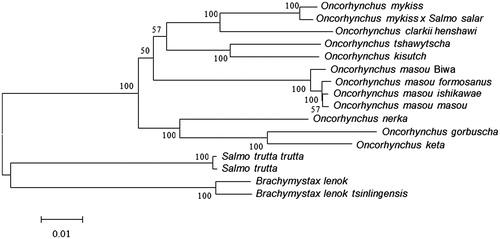
Multiple alignments of 16 mitochondrial genomes were performed using CLC Genomics Workbench with progressive alignment algorithm (Feng & Doolittle Citation1987). The phylogenetic tree from maximum-likelihood was reconstructed using MEGA6 (Tamura et al. Citation2013) (). To obtain the confident supports, one thousand bootstrap replicates were set for the analysis. Phylogenetic analysis showed that O. m. formosanus shared the same cluster with other O. masou subspecies. Oncorhynchus m. formosanus may diverge earlier than O. masou ishikawae and O. masou masou but later than O. masou Biwa. We expect that the present result will help to elucidate the taxonomic status of O. m. formosanus and contribute to the conservation.
Disclosure statement
The authors declare no competing interests in the preparation and execution of this manuscript. The authors are solely responsible for its content.
References
- Chung LC, Lin HJ, Yo SP, Tzeng CS, Yeh CH, Yang CH. 2008. Relationship between the Formosan landlocked salmon Oncorhynchus masou formosanus population and the physical substrate of its habitat after partial dam removal from Kaoshan Stream, Taiwan. Zool Stud. 47:25–36.
- Feng DF, Doolittle RF. 1987. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 25:351–360.
- Gwo JC, Hsu TH, Lin KH, Chou YC. 2008. Genetic relationship among four subspecies of cherry salmon (Oncorhynchus masou) inferred using AFLP. Mol Phylogenet Evol. 48:776–781.
- Hsu CB, Tzeng CS, Yeh CH, Kuan WH, Kuo MH, Lin HJ. 2010. Habitat use by the Formosan landlocked salmon Oncorhynchus masou formosanus. Aquatic Biol. 10:227–239.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Jordan DS, Oshima M 1919. Salmo formosanus, a new trout from the mountain streams of Formosa. Proc Acad Nat Sci Philadelphia. 71:122–124.
- Kottelat M. 1996. Oncorhynchus formosanus. The IUCN Red List of Threatened Species 1996: e.T15323A4513507; [cited 2016 Mar 21]. Available from: http://dx.doi.org/10.2305/IUCN.UK.1996.RLTS.T15323A4513507.en.
- Numachi K, Kobayashi T, Chang K, Lin Y. 1990. Genetic identification and differentiation of the Formosan landlocked salmon, Oncorhynchus masou formosanus by restriction analysis of mitochondrial DNA. Bull Inst Zool Acad Sin. 29:61–72.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Yang JQ, Cheng HL, Wu CY, Tsai KC, Chiang HC, Weng CW, Lin HD. 2013. Complete mitochondrial genome of Scaphiodonichthys acanthopterus (Cypriniformes, Cyprinidae). Mitochondrial DNA. 24:108–109.