Abstract
In this study, the mitochondrial genome of Melanotaenia australis Castelnau, 1875 (Atheriniformes:Melanotaeniidae) was sequenced for the first time. The assembled mitogenome consisting of 16,530 bp, includes 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs genes and one putative control region. The overall base composition of M. australis is 27.79% for A, 29.66% for C, 15.90% for G, 26.66% for T and shows 92% identities to Lake Kutubu Rainbowfish, Melanotaenia lacustris. These data would provide useful molecular information for phylogenetic relationships within the family Melanotaeniidae species.
The Melanotaenia australis (Castelnau, 1875) is one of the most common and widespread freshwater fish in river systems of north-western Australia. Melanotaenia australis can grow to a length of around 10 cm, but are more common at 8 cm or less. Males are usually much larger and deeper bodied than females. Colouration generally consists of 1–2 broad, dark mid-lateral stripes and a series of narrow longitudinal stripes of orange-red corresponding with each scale row. The whole body has an overall sheen of iridescent purple or blue. Fins range from nearly colourless to deep red, or clear with red or green flecks. The second dorsal and anal fins have reddish orange rays with a yellow membrane and dark distal border (Allen Citation1978).
Samples of M. australis were wild captured from a river of the Kimberley region (north-western Australia), then the muscle was dissected and preserved in pure alcohol. The specimens were stored in Fish Specimens Museum in Shanghai Ocean University, the accession number is SHOU20150061001. Then their genomic DNA was extracted from muscle by using Sangon Mag-MK Animal Genomic DNA extraction kit (Sangon, Shanghai, China). The primers were designed according to the complete mitochondrial genome of Melanotaenia boesemani (KT380951) deposited in GenBank. PCR amplification and sequencing of the products were performed according to the method described by He et al. (Citation2011) with slight modifications.
The complete mitochondrial genome of M. australis was 16,530 bp in size (GenBank accession no. KU529305), including 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs genes and one putative control region. All protein-coding genes are encoded on H-strand with exception of protein-coding genes of ND6. All tRNA genes are encoded on H-strand with exception of tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser (UGA), tRNA-Glu and tRNA-Pro. All the 13 mitochondrial protein-coding genes share the start codon ATG, except for COI (GTG start codon). The stop codon, TAA, is present in COI, ND4L and ND6; TAG is present in ND1, ATP8, ND5 and Cytb; an incomplete stop codon ‘‘TA–’’ is found in ND2, ATP6 and COIII; and ‘‘T– –’’ is found in COII, ND3 and ND4. The longest one is ND5 gene (1839 bp) in all protein-coding genes, whereas the shortest is ATP8 gene (168 bp). The size of the 22 tRNA ranges from 67 bp to 74 bp. The two ribosomal RNA genes, 12S rRNA gene (945 bp) and 16S rRNA gene (1677 bp), are located between tRNA-Phe and tRNA-Leu (UAA) and separated by tRNA-Val.
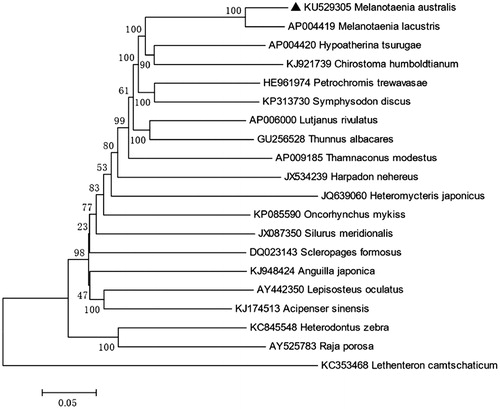
The control region, which is 873 bp, is located between tRNA-Pro and tRNA-Phe. The overall base composition of M. australis is 27.79% for A, 29.66% for C, 15.90% for G, 26.66% for T, which is characteristic of mitochondrial genomes of other bony fish (Miya et al. Citation2003; Zhao et al. Citation2015a,Citationb), and shows 92% identities to Lake Kutubu Rainbowfish, M. lacustris (). We expect that the present result would elucidate the further phylogenetic approach among different species of rainbowfishes.
Disclosure statement
The authors report that they have no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This work was supported by a grant from Shanghai Collaborative Innovation Center for Aquatic Animal Genetics and Breeding (ZF1206).
References
- Allen GR. 1978. The rainbow fishes of northwestern Australia (Family Melanotaeniidae). Tropical Fish Hobbyist. 26:91–102.
- He A, Luo Y, Yang H, Liu L, Li S, Wang C. 2011. Complete mitochondrial DNA sequences of the Nile tilapia (Oreochromis niloticus) and Blue tilapia (Oreochromis aureus): genome characterization and phylogeny applications. Mol Biol Rep. 38:2015–2021.
- Miya M, Takeshima H, Endo H, Ishiguro NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, et al. 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol. 26:121–138.
- Zhao Y, Chen Z, Gao J, Wang L, Li Z. 2015a. The complete mitochondrial genome of boeseman's rainbowfish (Melanotaenia boesemani Allen & Cross, 1980). Mitochondrial DNA. 2015:1–2. DOI:10.3109/19401736.2015.1089556
- Zhao Y, Chen Z, Gao J, Wang L, Li Z, Yu Y, Zhou Q. 2015b. The complete mitochondrial genome of red rainbowfish (Glossolepis incises Weber 1907). Mitochondrial DNA. 2015:1–2. DOI:10.3109/19401736.2015.1079882