Abstract
The complete sequence of the mitochondrial genome of the Culex tritaeniorhynchus (Diptera: Culicidae) is presented using traditional Sanger sequencing. Its mitogenome are 15,123 bp in length, consisting of 13 protein-coding genes, 2 ribosomal RNA genes (rRNA) and 22 transfer RNA (tRNA) genes. The percent of A + T of the mitogenome is 77.73%. Most mitochondrial genes are encoded on the heavy strand, except for NAD5, NaD4, NAD4L, NAD1, two rRNA and nine tRNA genes, which are encoded on the light strand. The results of phylogenetic analyzes showed that the Cx. tritaeniorhynchus has close relationship with the species of genus Culex.
Culex tritaeniorhynchus (Diptera: Culicidae) is the most important vector of Japanese Encephalitis virus. It even can be superinfected with different viruses (Kuwata et al. Citation2015). ND4 and ND5 sequences of mosquito mitochondrion can provide evidence of population genetic structure and gene isolation by distance (Gorrochotegui-Escalante et al. Citation2000; Birungi & Munstermann Citation2002; Szalanski et al. Citation2006; Venkatesan et al. Citation2007). Cytochrome C oxidase I (COI) gene in mosquito mitochondrion is a very important target for identification DNA barcodes (Kumar et al. Citation2007; Wang et al. Citation2012; Ondrejicka et al. Citation2014). We first determined the mitochondrial genome of Cx. tritaeniorhynchus (GenBank accession no. KT852976). The sample were collected in Nanjing, China (geographic location: 118.84159°E, 32.01337°N) in August 2014. The collected specimen is stored in the Medical Insect Collection of Beijing Institute of Microbiology and Epidemiology, and its accession number is ME-Z-III-26-08.
The complete mitogenome of this individual is 15,123 bp in length, and consists of 13 protein-coding genes, two ribosomal RNA genes (rRNA) and 22 transfer RNA (tRNA) genes. The percent of A + T of the mitogenome is 77.73%. Mitochondrial DNA is circular double strands including heavy strand and light strand. Most mitochondrial genes are encoded on the heavy strand, except for NAD5, NAD4, NAD4L, NAD1, two rRNA and nine tRNA genes, which are encoded on the light strand. There are six mitochondrial protein-coding genes share the start codon ATG which are COX2, ATP6, COX3, NAD4, NAD4L and COB. The ATP8, NAD1, NAD6 and NAD6 are initiated with the start codon ATA. Both NAD2 and NAD5 are initiated with TCG. And the COX1 is initiated by TCG. The COX1, COX2 and NAD6 are terminated by TAA and AAA, respectively. The remaining 12 protein-coding genes end with the typical termination codon TAA. It is expected that this complete mitogenome could be used to investigate the phylogenetic relationship of Cx. tritaeniorhynchus in Culicidae and provide other more information.
In order to explore the evolution situation of Cx. tritaeniorhynchus in Diptera, we analyze the molecular phylogenetics of Diptera using the complete mitochondrial genome sequence of 18 species which are downloaded from the GenBank (). The 18 species come from three families: Culicidae, Muscidae and Drosophilidae.
Figure 1. Phylogenetic tree generated using the neighbour-joining method based on complete mitochondrial genomes. Aedes aegypti (EU352212), Ae. albopictus (KR068634), Ae. notoscriptus (KM676219), Ae. vigilax (KP995260), Culex tritaeniorhynchus (KT852976), Cx. pipiens pipiens (HQ724614), Cx. quinquefasciatus (HQ724617), Anopheles alitarsis (HQ335349), An. janconnae (HQ335348), An. quadrimaculatus (L04272), An. cracens (JX219733), An. gambiae (L20934), Musca domestica (KM200723), Stomoxys calcitrans (DQ533708), Drosophila simulans (AF200838), Dr. santomea (KF824871) and Dr. yakuba (KF824893).
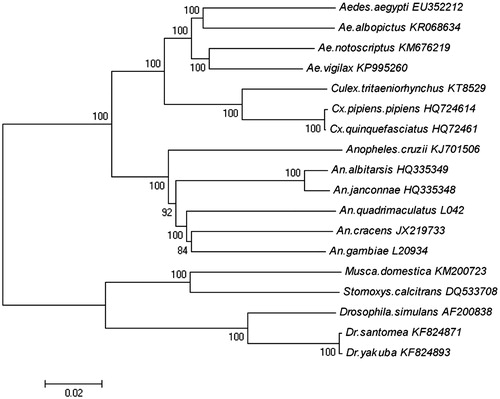
The mitochondrial genome was analyzed with neighbour-joining method, and the phylogenetic tree obtained is shown in . It shows that the three families in Diptera are phyletic lineages. In the three families, Culicidae comprised Aedes aegypti (EU352212), Ae. albopictus (KR068634), Ae. notoscriptus (KM676219), Ae. vigilax (KP995260), Culex tritaeniorhynchus (KT852976), Cx. pipiens pipiens (HQ724614), Cx. quinquefasciatus (HQ724617), Anopheles alitarsis (HQ335349), An. janconnae (HQ335348), An. quadrimaculatus (L04272), An. cracens (JX219733), An. gambiae (L20934); Mucidae comprised Musca domestica (KM200723); Stomoxys calcitrans (DQ533708). Drosophilidae comprised Drosophila simulans (AF200838), Dr. santomea (KF824871), Dr. yakuba (KF824893).
Funding information
This work was funded by grants from Infective Disease Prevention and Cure Project of China (No. 2012ZX10004219).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Birungi J, Munstermann LE. 2002. Genetic structure of Aedes albopictus (Diptera: Culicidae) population based on mitochondrial ND5 sequence: evidence for an independent invasion into Brazil and United States. Ann Entomol Soc Am. 95: 125–132.
- Gorrochotegui-Escalante N, Munoz ML, Femandes-Salas I, Beaty BJ, Black WC. 2000. Genetic isolation by distance among Aedes aegypti populations along the northeastern coast of Mexico. AM J Trop Med Hyg. 62:200–209.
- Kumar NP, Rajavel AR, Natarajan R, Jambulingam P. 2007. DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae). J Med Entomol. 44:1–7.
- Kuwata R, Isawa H, Sasaki T, Kobayashi M, Maedae K, Sawabe K. 2015. Analysis of mosquito-borne flavivirus superinfection in Culex tritaeniorhynchus (Diptera: Culicidae) cells persistently infected with culex flavivirus (Flaviviridae). J Med Entomol. 52:222–229.
- Ondrejicka DA, Locke SA, Morey K, Borisenko AV, Hanner RH. 2014. Status and prospects of DNA barcoding in medically important parasites and vectors. Trends Parasitol. 30:582–591.
- Szalanski AL, Owens CB, Lewter JA, Broce AB. 2006. Genetic structure of Aedes vexans (Diptera: Culicidae) populations from central United States based on mitochondrial ND5 sequences. Ann Entomol Soc Am. 99:157–163.
- Venkatesan M, Westbrook CJ, Hauer MC, Rasgon JL. 2007. Evidence for a population expansion in the West Nile virus vector Culex tarsalis. Mol Biol Evol. 24:1208–1218.
- Wang G, Li C, Guo X, Xing D, Dong Y, Wang Z, Zhang Y, Liu M, Zheng Z, Zhang H, et al. 2012. Identifying the main mosquito species in China Based on DNA barcoding. Plos One. 7:e47051.
