Abstract
Lilium tsingtauense, known as ‘Korean wheel lily’ and ‘Twilight lily’, is a lily species naturally distributed in Korea. The complete chloroplast genome sequences of L. tsingtauense was obtained by de novo assembly using whole-genome next-generation sequencing data. The chloroplast genome of L. tsingtauense was 152,710 bp in length and consisted of four distinct regions, such as large single-copy region (82,059 bp), small single-copy region (17,619 bp) and a pair of inverted repeat regions (26,516 bp). The genome contained a total of 113 genes, including 79 protein-coding genes, 30 tRNA genes and 4 rRNA genes. Phylogenetic analysis with the reported chloroplast genomes revealed that L. tsingtauense is most closely related to Lilium hansonii, a lily species native to Korea.
Lilium tsingtauense (also known as ‘Korean wheel lily’ and ‘Twilight lily’) is a lily species belonging to the Liliaceae family and naturally distributed in the Korean Peninsula and the Shandong Peninsula (Liang & Tamura Citation2000; Fox Citation2006; Lim et al. Citation2014; Park et al. Citation2014). L. tsingtauense shows unique flower morphology different from those of Lilium distichum and Lilium hansonii (Liang & Tamura Citation2000; Du et al. Citation2014). Although several phylogenetic studies reported nuclear and chloroplast sequences in L. tsingtauense (Nishikawa et al. Citation1999; Hayashi & Kawano Citation2000; Lee et al. Citation2011; Chen et al. Citation2013; Gao et al. Citation2013), the complete chloroplast genome sequence is not available for this plant species. We report here the complete chloroplast genome sequence of L. tsingtauense and phylogenetic relationship of this plant with other neighbour species in the Liliaceae family.
Total genomic DNAs were purified from mature leaves sampled from a natural habitat in Mt. Chogae (35°0′ 9.33" N, 127°18′ 48.79" E), Sunchun, South Korea (Kyungpook National University voucher no. KNU IT No. 121015-1) using a modified CTAB method (Allen et al. Citation2006). An illumina paired-end (PE) genomic library was constructed and sequenced using an Illumina HiSeq platform, according to standard Illumina PE protocol. PE reads of 1.9 Gb were obtained (SRA accession no. SRX529350) and assembled by a CLC genome assembler (ver. 4.06 beta, CLC Inc, Aarhus, Denmark), as mentioned in Kim et al (Citation2015b, Citation2015c). The representative chloroplast contigs were retrieved, ordered and joined into a single draft sequence, by comparison with the chloroplast sequence of Lilium longiflorum (KC968977) as a reference. The draft sequence was examined and manually corrected by PE read mapping. The genes in the chloroplast genome were predicted using the DOGMA program (Wyman et al. Citation2004) and BLAST searches.
The complete chloroplast genome of L. tsingtauense (GenBank accession KM103365) was 152,710 bp long and consisted of four distinct regions: large single-copy (LSC) region of 82,059 bp, small single-copy (SSC) region of 17,619 bp and a pair of inverted repeat regions of 26,516 bp. Overall, GC contents of chloroplast genomes was 37.0%. The chloroplast genome contained a total of 113 genes, including 79 protein-coding genes, 30 tRNA genes and 4 rRNA genes.
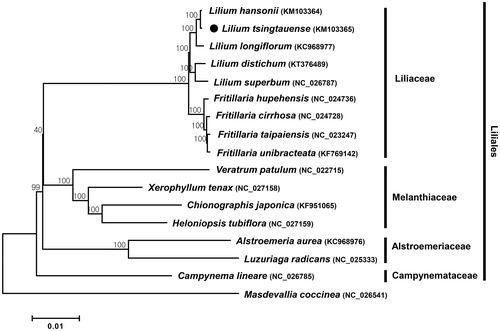
Phylogenetic analysis was carried out using 79 protein-coding sequences of L. tsingtauense with those of 15 species in the Liliales order by a neighbour-joining (NJ) analysis using MEGA 6.0 (Tamura et al. Citation2013) (). The phylogenetic tree provides four independent groups according the family to where species belong, as described in Hwang et al (Citation2016). In the tree, L. tsingtauense was subgrouped with other Lilium species within the Liliaceae family and placed more closely to L. hansonii, a lily species native to Korea (Kim et al. Citation2015a).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This work was carried out with the support of ‘Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01095307)’ Rural Development Administration, Republic of Korea.
References
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1:2320–2325.
- Chen S, Kim DK, Chase MW, Kim JH. 2013. Networks in a large-scale phylogenetic analysis: reconstructing evolutionary history of Asparagales (Lilianae) based on four plastid genes. PLoS One. 8:e59472.
- Du YP, He HB, Wang Zx Wei C, Li S, Jia GX. 2014. Investigation and evaluation of the genus Lilium resources native to China. Genet Resour Crop E. 61:395–412.
- Fox EE. 2006. Martagon lilies: old world, whorled-leaf lilies. Millet, Alberta, Canada.
- Gao YD, Harris AJ, Zhou SD, He XJ. 2013. Evolutionary events in Lilium (including Nomocharis, Liliaceae) are temporally correlated with orogenies of the Q-T plateau and the Hengduan Mountains Mol Phylogenet Evol. 68:443--460.
- Hayashi K, Kawano S. 2000. Molecular systematics of Lilium and allied genera (Liliaceae): phylogenetic relationships among Lilium and related genera based on the rbcL and matK gene sequence data. Plant Spec Biol. 15:73–93.
- Hwang YJ, Lee SC, Kim K, Choi BS, Park JY, Yang TJ, Lim KB. 2016. The complete chloroplast genome of Lilium distichum Nakai (Liliaceae). Mitochondrial DNA Part A. Published online: 09 Feb 2016. DOI:10.3109/19401736.2015.1101591.
- Kim K, Hwang YJ, Lee SC, Yang TJ, Lim KB. 2015a. The complete chloroplast genome sequence of Lilium hansonii Leichtlin ex D.D.T.Moore. Mitochondrial DNA. Early Online 1–2. DOI: 10.3109/19401736.2015.1079852.
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ. 2015b. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One 10:e0117159.
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015c. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Lee CS, Kim SC, Yeau SH, Lee NS. 2011. Major lineages of the genus Lilium (Liliaceae) based on nrDNA its sequences, with special emphasis on the Korean species. J Plant Biol. 54:159–171.
- Liang S, Tamura MN. 2000. Lilium. In: Wu ZW, Raven PH, editors. Flora of China Liliaceae, Vol. 24. Beijing, China and St. Louis, Missouri, (MO), USA: Science Press and Missouri Botanical Garden Press; [cited 2016 March 23]. Available from: http://www.efloras.org/florataxon.aspx?flora_id =2&taxon_id =118558.
- Lim KB, Younis A, Park JT, Hwang YJ. 2014. Exploitation of diversity for morphological traits in Lilium tsingtauense under different habitats. Not Sci Biol. 6:178–184.
- Nishikawa T, Okazaki K, Uchino T, Arakawa K, Nagamine T. 1999. A molecular phylogeny of Lilium in the internal transcribed spacer region of nuclear ribosomal DNA. J Mol Evol. 49:238–249.
- Park JT, Hwang YJ, Lee HI, Younis A, Lim KB. 2014. Ecological analysis of Lilium tsingtauense native in Korea. Hort Environ Biotechnol. 55:230–236.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.