Abstract
The complete mitochondrial genome of the Lake Baikal sponge Baikalospongia intermedia was sequenced. The circular mitochondrial genome is 28,327 bp in length and includes 14 protein-coding genes, 2 ribosomal RNA genes and 25 transfer RNA genes. Bayesian comparative analysis of molecular evolution rates was found no acceleration of the mtDNA evolution of B. intermedia. This species clustered with other species of the genus Baikalospongia on the Bayesian tree.
Keywords:
Sponges of Lake Baikal (Lubomirskiidae) are an endemic group of freshwater sponges (Demospongiae, Haplosclerida) that originated from the cosmopolitan family Spongillidae (Itskovich et al. Citation2008) during last ca. 10 MYA according to the molecular phylogeny (Maikova et al. Citation2015). Phylogenetic relationships within the family Lubomirskiidae are still poorly understood (Efremova Citation2001; Itskovich et al. Citation2006, Citation2008). Earlier, in order to reveal phylogenetic relationships, sequences of six mitochondrial genomes of Baikal sponges were determined (Lavrov Citation2010; Lavrov et al. Citation2012; Maikova et al. 2015). Here, we described a new mtDNA of Baikalospongia intermedia, which will be a useful instrument for future phylogenetic analyses of Baikal endemic sponges.
B. intermedia is one of the most abundant species in Lake Baikal, which is distributed widely in the lake at the depth range of 3 to 40 m (Efremova Citation2001). A specimen was collected from a depth of 20 m near the village of Bolshiye Koty (51°54" 16.66" N, 105°5′ 28.36" E) in August 2008 and kept under number BS121 in the sponge collection at Laboratory of Analytical and Bioorganic Chemistry of Limnological Institute (Irkutsk, Russia).
The species was identified from spicules and skeleton of the sponge. Spicule and skeleton preparations were performed as previously described (Efremova Citation2001). Total DNA was extracted from tissue by a modified phenol–chloroform method (Maniatis et al. Citation1984). Genome was amplified and sequenced as previously described (Maikova et. al. 2015). The genome was assembled using MAFFT v 6.882 (Katoh & Toh Citation2008). In mitochondrial DNA sequence of B. intermedia, it was problematic to sequence a short fragment within the gene for small subunit ribosomal RNA (rns) and a section of the intergenic region (IGR) between trnQ and trnN genes, including trnW gene, because of probable additional hairpin structures. Expected total length of problematic regions is 1186 bp based on the alignment to the closely related mitochondrial genome of L. baicalensis. Annotated mitochondrial genome sequence of B. intermedia is available in NCBI (GenBank accession number KU324767).
The mtDNA of B. intermedia is 28,327 bp and contains 2 rRNA genes, 25 tRNA genes and 14 protein-coding genes. Total length of intergenic regions is 9136 bp, which constitutes 32% of the genome. The A + T base composition of the genome is 59.05%, the A + T content of genes ranging from 48.86% to 68.94% ().
Table 1. The genomic organization of mitochondrial genes of Baikalospongia intermedia.
The genome contains tRNA genes for all amino acids. However, the anticodons encoded match only 39% of all codons in the genome. For example, tRNAIle with codon AUC matching GAU is only 8.7% of all codons coding this amino acid. Supposedly, at least some synonymic mutations may be nonneutral in case if codon composition affects the translation rate of the corresponding gene (Zhou et al. Citation2013).
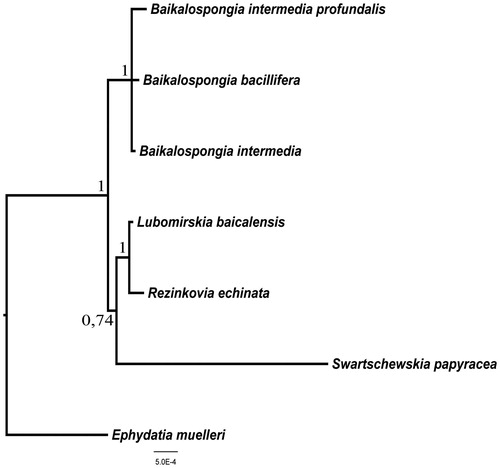
Bayesian comparative analysis (Ronquist & Huelsenbeck Citation2003) found no significant difference between mitochondrial DNA substitution rate of B. intermedia and other Baikal sponges (95% HPD interval <1, variance >1). B. intermedia clustered on the phylogenetic tree with other species of the genus Baikalospongia ().
Figure 1. Bayesian tree inferred from coding sequences of all 14 mitochondrial genes of six species from Lubomirskiidae (R. echinata (JQ302309), S. papyracea (JQ302308), B. intermedia profundalis (JQ302310), L. baicalensis (GU385217), B. bacillifera (KJ192328), B. intermedia (KU324767)) and one species from Spongillidae (E. muelleri (NC_010202)) as out group. For Bayesian analyses, the Markov chain Monte Carlo search was run twice (default parameter) on four chains for 5,000,000 generations/trees were sampled every 1000th cycle after the first 10,000 burn-in cycles. Values above and to the left of nodes are Bayesian posterior probabilities.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding
This work was supported by the RFBR grant No. 14-04-31298 and the governmentally funded project No. VI. 50. 1. 4.
References
- Efremova SM. 2001. Porifera. In: Timoshkin OA, editor. Index of animal species inhabiting Lake Baikal and its catchment area, Vol. 1. Novosibirsk: Nauka. p. 179–192.
- Itskovich V, Belikov S, Efremova S, Masuda Y. 2006. Monophyletic origin of freshwater sponges in ancient lakes based on partial structures of COXI gene. Hydrobiologia. 568:155–159.
- Itskovich V, Gontcharov A, Masuda Y, Nohno T, Belikov S, Efremova S, Meixner M, Janussen D. 2008. Ribosomal ITS sequences allow resolution of freshwater sponge phylogeny with alignments guided by secondary structure prediction. J Mol Evol. 167:608–620.
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 9:286–298.
- Lavrov DV. 2010. Rapid proliferation of repetitive palindromic elements in mtDNA of the endemic Baikalian sponge Lubomirskia baicalensis. Mol Biol Evol. 27:757–760.
- Lavrov DV, Maikova OO, Pett W, Belikov SI. 2012. Small inverted repeats drive mitochondrial genome evolution in Lake Baikal sponges. Gene. 505:91–99.
- Maikova O, Khanaev I, Belikov S, Sherbakov D. 2015. Two hypotheses of the evolution of endemic sponges in Lake Baikal (Lubomirskiidae). J Zoolog Syst Evol Res. 53:175–179.
- Maniatis T, Fritsch EF, Sambrook J. 1984. Molecular cloning. Moscow: Mir, 333–337. (Russian Translation).
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Zhou M, Guo J, Cha J, Chae M, Chen S, Barral JM, Sachs MS, Liu SYi. 2013. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature. 495:111–115.
