Abstract
The complete mitogenome genome of Calliphora chinghaiensis (Diptera: Calliphoridae) is determined in this study. Mitochondrion of C. chinghaiensis is 15,269 bp long. It forms a circular DNA molecule that includes 37 typical animal mitochondrial genes and an A + T-rich region. All protein-coding genes are initiated by ATN start codon, except for cox1, which uses TCG as its start codon. 11 protein-coding genes stop with the termination codon TAN, while other protein-coding genes use CTT and AGT (cytb) as termination codon, respectively. Furthermore, the largest non-coding A + T-rich region with a length of 442 bp is at the end of rrns. The mitochondrial genome of C. chinghaiensis has been completely sequenced for the first time in this study.
Mitochondrial DNA (mtDNA) has been widely used in phylogenetic research studies, phylogeography, population structure and dynamics, and molecular evolution (Boore Citation1999; Shi et al. Citation2015). There is no complete mitochondrial genome of Calliphora chinghaiensis (Diptera: Calliphoridae) has been reported by far. In 1926, the mitochondrial genome of C. chinghaiensis was primitively named. Usually, C. chinghaiensis inhabited in mountains over 3000 m and distributed at Qinghai, Sichuan, Yunnan and Tibet in China (Hu et al. Citation2007). Specimens of C. chinghaiensis (specimen voucher SIE32066838) were collected form Shangri-la, Yunnan Province, China (N: 26°–34°, E: 94°–102°). They were identified as C. chinghaiensis through external morphology. Our DNA extraction method followed direction of DNA extraction kit by DNeasy Blood & Tissue Kit (Sangon Biotech, China). The complete mitochondrial genome of C. chinghaiensis (Diptera: Calliphoridae) has been submitted to GenBank under accession no. KT936147.
The length of complete mitochondrial genome of C. chinghaiensis is 15,269 base pairs (bp), with the overall A + T content of 76.73% (A = 39.25%, G = 9.95%, T = 37.48% and C = 13.28%). The genome encodes the same set of 37 genes including 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs) and two ribosomal RNAs (rRNAs) (Clary & Wolstenholme Citation1987). In addition, the complete mitochondrial genome contains a non-coding A + T-region (D-loop) related to the regulation of transcription and the control of replication (Chang & Clayton, Citation1984; Shadel & Clayton, Citation1993). All the PCGs initiate with an ATN start codon (nd2, nd3, nd5 nd6 and cox3 start with ATT; nd4, nd4l, cox2, cytb and atp6 start with ATG; atp8 starts with ATC and nd1 starts with ATA) except for cox1, which initiates with TCG start codon. The TCG as a serine is assigned as the cox1 start codon, since the hexanucleotide ATTTAA participating initiation signalling is found next to a TCG codon (Junqueira et al. Citation2004). Therefore, the TCG for a serine has been assigned as the cox1 start codon. The hexanucleotide signals were detected in mosquitoes (Beard et al. Citation1993; Mitchell et al. Citation1993). Eight PCGs (nd1, nd2, nd3, nd4l, nd6, cox1, cox3 and atp8) contain the typical termination codon TAA, one gene (cytb) stops with AGT, one gene (cox2) terminates with CTT. While nd4, nd5 and atp6 have a stop codon of TAT.
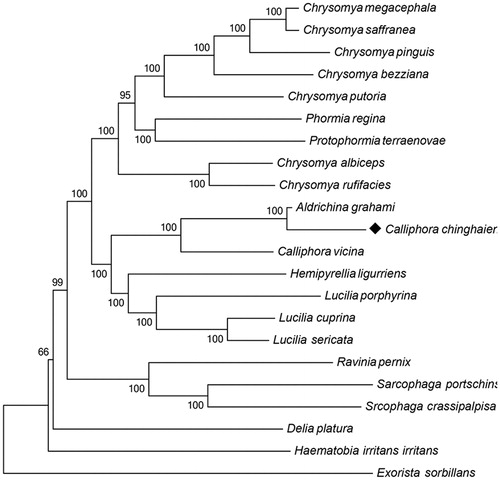
Like other insect tRNAs, the mitochondrial genome of C. chinghaiensis contains 22 transfer RNA genes, ranging from 64 to 72 bp, has a typical cloverleaf structure. The large mitochondrial rRNA subunit (rrnL) is 1328 bp long with an A + T content of 81.93%, while the small subunit (rrnS) has a total length of 785 bp, with an A + T content of 78.34%. The largest non-coding A + T-rich region is 442 bp long, which is shorter than the rrnS (785 bp). The A + T content is up to 84.16%, the highest of any region in the whole genome was believed to participated in the regulation of transcription and control of DNA replication (Zhang & Hewitt Citation1997). A neighbour-joining (NJ) tree of 16 Calliphoridae and six outgroups (R. pernix, D. platura, H. irritans irritans, S. portschinskyi, S. crassipalpisa, and E. sorbillans) based on 13 protein-coding genes is constructed by MEGA6 (Tamura et al. Citation2013) with the Poisson model (). The NJ bootstrap for 1000 replicates was indicated in each node. The evolutionary position of C. chinghaiensis was showed in the NJ tree. Except C. chinghaiensis, all other mitochondrial genomes were obtained from NCBI.
Figure 1. Evolutionary relationships of 16 Calliphoridae and 6 outgroup species. The mitochondrial genome of this study has been marked as black. GenBank IDs: A. grahami, KP872701.1; C. albiceps, JX913736.1; C. bezziana, JX913737.1; C. chinghaiensis, KT936147; C. putoria, AF352790.1; C. vicina, JX913760.1; C. megacephala, JX913739.1; C. saffranea, JX913742.1; C. pinguis, KM244730.1; C. rufifacies, JX913740.1; H. ligurriens, JX913759.1; P. regina, KC005712.1; P. terraenovae, JX913743.1; L. sericata, JX913756.1; L. cuprina, JX913749.1; L. porphyrina, JX913758.1; R. pernix, KM676414.1; D. platura, KP901268.1; H. irritans irritans, DQ029097.1; S. portschinskyi, KM287570.1; S. crassipalpisa, KP861920.1; E. sorbillans, HQ322500.1.
Funding information
This work was supported by the Program from Sichuan Agricultural University (02920400); National Natural Science Foundation of China (NSFC31160432) and Sichuan Provincial Department of Science and Technology Program (2015JQO023).
Disclosure statement
The authors declare no competing interests.
References
- Beard C, Hamm D, Collins F. 1993. The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol Biol. 2:103–124.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Chang DD, Clayton DA. 1984. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 36:635–643.
- Clary DO, Wolstenholme DR. 1987. Drosophila mitochondrial DNA: conserved sequences in the A + T-rich region and supporting evidence for a secondary structure model of the small ribosomal RNA. J Mol E. 25:116–125.
- Hu A, Ming-Fu W, Yang X. 2007. Preliminary studies on diversity of C alliphoridae (Diptera: Calliphoridae) in the Qinghai-Tibetan Plateau. Entomol J East China. 16:285–289.
- Junqueira AC, Lessinger AC, Torres TT, Da Silva FR, Vettore AL, Arruda P, Azeredo Espin AM. 2004. The mitochondrial genome of the blowfly Chrysomya chloropyga (Diptera: Calliphoridae). Gene. 339:7–15.
- Mitchell SE, Cockburn AF, Seawright JA. 1993. The mitochondrial genome of Anopheles quadrimaculatus species A: complete nucleotide sequence and gene organization. Genome 36:1058–1073.
- Shadel GS, Clayton DA. 1993. Mitochondrial transcription initiation. Variation and conservation. J Biol Chem. 268:16083.
- Shi X, Zhu Q, Wu N, Tu J, Yang D, Xu H, Yao Y, Yang M, Li D. 2015. The complete nucleotide sequence of the mitochondrial genome of Drosophila formosana (Diptera: Drosophilidae). Mitochondrial DNA. [Epub ahead of print]. doi:10.3109/19401736.2015.1089480.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zhang DX, Hewitt GM. 1997. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem System Ecol. 25:99–120.
