Abstract
Three complete mitochondrial genomes of South American electric fishes (Gymnotiformes), derived from high-throughput RNA sequencing (RNA-Seq), are reported herein. We report the complete mitochondrial genome of the bluntnose knifefish Brachyhypopomus n.sp. VERD, determined from newly sequenced data. We also provide the complete mitochondrial genomes for Sternopygus arenatus and the electric eel Electrophorus electricus, assembled from previously published transcriptome data. The mitochondrial genomes of Brachyhypopomus n.sp. VERD, Sternopygus arenatus and Electrophorus electricus have 13 protein-coding genes, 1 D-loop, 2 ribosomal RNAs and 22 transfer RNAs, and are 16,547, 16,667 and 16,906 bp in length, respectively. Phylogenetic analysis of the eight available mitochondrial genomes of gymnotiform fishes shows Apteronotus to be the sister lineage of other gymnotiformes, contradicting the “Sinusoidea” hypothesis that Apteronotidae and Sternopygidae are sister taxa.
Keywords:
The Gymnotiformes are a diverse group of more than 200 species of neotropical ostariophysan fishes that produce electric signals for both navigation and communication. The species of this order have been used as models in neuroscience, including studies of electrosensory modulation (Márquez et al. Citation2013) and central nervous system regeneration (Zupanc & Sîrbulescu Citation2013). To date, complete mitochondrial genomes have been reported for the gymnotiforms Apteronotus albifrons, Brachyhypopomus occidentalis, Gymnorhamphichthys sp. and Eigenmannia sp., and near-complete mitochondrial genomes have been reported for Gymnotus carapo and Electrophorus electricus (Nakatani et al. Citation2011). Here, we report the complete mitochondrial genome for the undescribed species Brachyhypopomus n.sp. VERD (Crampton et al. unpublished) which we assembled from newly generated RNA-seq data. We also report the complete mitochondrial genomes for Sternopygus arenatus and Electrophorus electricus, based on assemblies of RNA-seq data produced by Gallant et al. (Citation2014).
Electric organ was collected from a fresh specimen of Brachyhypopomus n.sp. VERD in the field (voucher specimen MUSM 54490, 04°53.986′ S, 073°38.849′ W) and preserved in RNAlater (Qiagen, Hilden, Germany). The electric organ was homogenized in Trizol (Invitrogen, Carlsbad, CA) using a BeadBug (Benchmark Scientific, Edison, NJ). Total RNA was extracted following a combined Trizol/RNeasy (Qiagen) protocol according to the manufacturer’s instructions. Library construction and sequencing on the Illumina HiSeq pipeline were performed at The Centre for Applied Genomics, the Hospital for Sick Children (Toronto). Sternopygus and Electrophorus raw reads were obtained from the NCBI SRA repository (SRR1299088, SRR1299078 and SRR1299082). For all datasets, sequencing reads were trimmed with Trimmomatic v0.33 (Bolger et al. Citation2014) using default settings. Trimmed reads were mapped to the phylogenetically closest mitochondrial genome currently available on Genbank using the map-to-reference feature of Geneious v6 (Biomatters, Aukland, New Zealand; Kearse et al. Citation2012): Brachyhypopomus n.sp. VERD reads were mapped to B. occidentalis (Genbank AP011570), Sternopygus arenatus to Eigenmannia sp. (Genbank AB054131) and Electrophorus electricus to the near-complete mitochondrial genome of E. electricus (Genbank AP011978.1). Consensus sequences of aligned reads were generated using the highest quality base calls. The assembled mitochondrial genomes were annotated using MitoAnnotator (Iwasaki et al. Citation2013).
To verify the species identities for both published and newly produced gymnotiform mitochondrial genomes, we compared the cytochrome b (cytb) and cytochrome oxidase I (coI) genes from the mitochondrial genomes to available sequences in Genbank, as well as a large database of cytb and coI sequences produced in the course of an ongoing multi-gene phylogenetic analysis of gymnotiform fishes (Janzen et al. unpublished). We confirmed the species identity of the mitochondrial genomes from Apteronotus albifrons, Brachyhypopomus occidentalis, Gymnotus carapo and Electrophorus electricus. We were unable to determine the species identity of Gymnorhamphichthys sp. or Eigenmannia sp. due to an absence of high sequence similarity matches with Genbank sequences or sequences in our database, but we did confirm the generic identity of these samples. Interestingly, our analysis suggests that the transcriptome reported from Sternopygus macrurus (SRR1299088) is actually from Sternopygus arenatus. The cytb and coI sequences from the mitochondrial genome assembled from this transcriptome data match S. arenatus with 99.8% sequence identity, but are only 90.6% identical to S. macurus. Phylogenetic analysis (not shown) using a dataset of Sternopygidae species (Maldonado-Ocampo Citation2011) indicate that the transcriptome data clusters with known S. arenatus sequences. Finally, we determined the nuclear zic1 sequence from the transcriptome and compared it to zic1 species from multiple sternopygid species, which confirmed the identity of this sample as S. arenatus.
The mitochondrial genomes of Brachyhypopomus n.sp. VERD (Genbank KX058570), Sternopygus arenatus (Genbank KX058571) and Electrophorus electricus (Genbank KX058572) are complete, with 13 protein-coding genes, 1 D-loop, 2 ribosomal RNAs and 22 transfer RNAs for a total length of 16,547, 16,667 and 16,906 bp, respectively. Base compositions are typical for vertebrate mitochondrial genomes with 28.7% A, 29.9% C, 15.7% G, 25.7% T for Brachyhypopomus n.sp. VERD, 31.3% A, 28.5% C, 14.6% G, 25.5% T for Sternopygus arenatus, and 31.3% A, 26.0% C, 13.6% G, 29.1% T, 39.6% for Electrophorus electricus.
To produce a phylogenetic tree using gymnotiform mitogenomes, we aligned the eight gymnotiform mitochondrial genomes with available Characiformes and Siluriformes and a cypriniform outgroup (Zacco platypus) using MAFFT (Katoh & Standley Citation2013). We then generated a maximum likelihood phylogeny using IQ-Tree (Nguyen et al. Citation2015) with 1000 bootstrap replicates (). We tested for the best-fit model based on Akaike Information Criterion (AIC), corrected Akaike Information Criterion (AICc) and Bayesian Information Criterion (BIC), which converged on the GTR + I + G4 model.
Figure 1. Phylogenetic relationships of gymnotiform electric knifefishes and allies based on maximum likelihood analysis of mitochondrial genomes. Numbers at nodes show bootstrap proportions. Mitochondrial genomes assembled for this study are in bold.
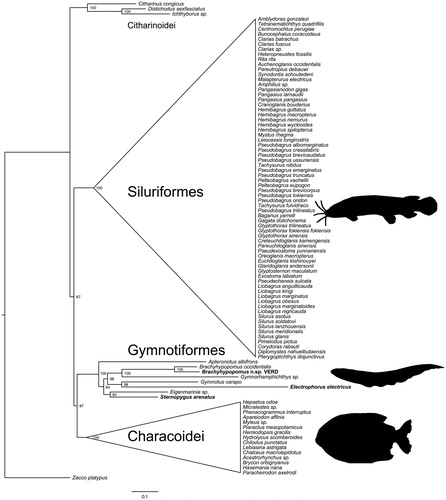
Our phylogenetic reconstruction confirms the monophyly of Siluriformes and Gymnotiformes, but indicates that Characiformes are non-monophyletic and are divided into Characoidei and Citharinoidei, as seen in other molecular studies (Nakatani et al. Citation2011; Chen et al. Citation2013), and in contrast to morphological evidence (see summary in Chen et al. Citation2013). Within Gymnotiformes, patterns of relationships largely match expectations based on taxonomy and prior phylogenetic studies. The two Brachyhypopomus species are sister taxa, and are most closely related to Gymnorhamphichthys, supporting a close relationship between the families Hypopomidae and Rhamphichthyidae (Maldonado-Ocampo et al. Citation2014). Gymnotus and Electrophorus are well-supported as sister taxa, corroborating the long-standing hypothesis that groups these genera in the family Gymnotidae (Albert & Crampton Citation2005; Lovejoy et al. Citation2010). Sternopygus and Eigenmannia, both members of the family Sternopygidae, group together as expected. The earliest diverging gymnotiform lineage has not been decisively determined. In this study, Apteronotus is sister to the other gymnotiform lineages, matching the pattern based on sodium channel sequences (Arnegard et al. Citation2010), but in contrast to other molecular and morphological studies that place Gymnotidae at the base of the gymnotiform tree (Albert & Crampton Citation2005; Tagliacollo et al. Citation2016). This topology contradicts a hypothesis that groups as sister taxa Sternopygidae and Apteronotidae, the families with wave-type electric organ discharges, in a “Sinusoidea” clade (Albert Citation2001).
The results reported here contribute to ongoing efforts to reconstruct teleost phylogenies based on mitochondrial genomes, and also highlight the value of mitochondrial genes as markers for confirming species identifications for genomic resources.
Funding information
This work was supported by National Sciences and Engineering Research Council (NSERC) Discovery grants (B.S.W.C. and N.R.L.), National Science Foundation (NSF) DEB-1146374 (W.G.R.C.), an Ontario Graduate Scholarship (R.K.S.), an Ontario Trillium Scholarship (M.A.K.) and Vision Science Research Program Scholarships (R.K.S and A.V.N.).
Acknowledgements
We thank Hernán Ortega, National University of San Marcos, Lima, Peru for assisting arrangement of fieldwork.
Disclosure statement
The authors do not report any conflict of interests, and are responsible for the content of this publication.
References
- Albert JS. 2001. Species diversity and phylogenetic systematics of American knifefishes (Gymnotiformes, Teleostei). Misc Pub Mus Zool Univ Mich 190:1–127.
- Albert JS, Crampton WGR. 2005. Diversity and phylogeny of Neotropical electric fishes (Gymnotiformes). In: Bullock TH, Hopkins CD, Popper AN, Fay RR, editors. Electroreception. New York: Springer. p. 360–409.
- Arnegard ME, Zwickl DJ, Lu Y, Zakon HH. 2010. Old gene duplication facilitates origin and diversification of an innovative communication system-twice. Proc Natl Acad Sci USA 107:22172–22177.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120.
- Chen WJ, Lavoué S, Mayden RL. 2013. Evolutionary origin and early biogeography of otophysan fishes (Ostariophysi: Teleostei). Evolution 67:2218–2239.
- Gallant JR, Traeger LL, Volkening JD, Moffett H, Chen PH, Novina CD, Phillips GN Jr, Anand R, Wells GB, Pinch M, et al. 2014. Genomic basis for the convergent evolution of electric organs. Science 344:1522–1525.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol 30:2531–2540.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649.
- Lovejoy NR, Lester K, Crampton WGR, Marques FP, Albert JS. 2010. Phylogeny, biogeography, and electric signal evolution of Neotropical knifefishes of the genus Gymnotus (Osteichthyes: Gymnotidae). Mol Phylog Evol 54:278–290.
- Maldonado-Ocampo J. 2011. Filogenia Molecular da Família Sternopygidae (Gymnotiformes: Sternopygoidei). PhD Thesis, Universidad Federal do Rio de Janeiro.
- Maldonado-Ocampo J, López-Fernández H, Taphorn DC, Bernard CR, Crampton WGR, Lovejoy NR. 2014. Akawaio penak, a new genus and species of neotropical electric fish (Gymnotiformes, Hypopomidae) endemic to the Upper Mazaruni River in the Guiana Shield. Zoologca Scripta 43:24–33.
- Márquez BT, Krahe R, Chacron MJ. 2013. Neuromodulation of early electrosensory processing in gymnotiform weakly electric fish. J Exp Biol 216:2442–2450.
- Nakatani M, Miya M, Mabuchi K, Saitoh K, Nishida M. 2011. Evolutionary history of Otophysi (Teleostei), a major clade of the modern freshwater fishes: Pangaean origin and Mesozoic radiation. BMC Evol Biol 11:177.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274.
- Tagliacollo VA, Bernt MJ, Craig JM, Oliveira C, Albert JS. 2016. Model-based total evidence phylogeny of Neotropical electric knifefishes (Teleostei, Gymnotiformes). Mol Phylog Evol 95:20–33.
- Zupanc GK, Sîrbulescu RF. 2013. Teleost fish as a model system to study successful regeneration of the central nervous system. In: Heber-Katz E, Stocum DL, editors. New perspectives in regeneration. Berlin: Springer. p. 193–233.
