Abstract
The whole complete chloroplast genome of Lagerstroemia macrocarpa was assembled in this study. Total genome is 152,472 bp in length consisting of two inverted repeats of 17,562 bp separated by a large single-copy region and a small single-copy region of 84,050 bp and 33,295 bp, respectively. This genome contains 112 unique genes including 78 protein-coding genes, 4 ribosomal RNA genes and 30 transfer RNA genes. In 78 protein-coding genes, 8 genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, rps16) contain one intron and three genes with two introns each (clpP, rps12 and ycf3). This newly sequenced chloroplast genome supply highly variable information of polymorphisms within Lagerstroemia species.
Lagerstroemia (‘crape myrtle’) is the most economically genus in Lythraceae and comprises about 55 species (Koehne Citation1883; Furtado & Srisuko Citation1969; Qin & Graham Citation2007) which mainly distributed in tropical and sub-tropical habitats of southern China, Japan and northeast Australia (Egolf & Andrick Citation1978). Most Lagerstroemia species blossom from summer till fall with good features of colorful flowers, being easily propagated, strongly resistant to multiple pathogens (Wang et al. Citation2011). In terms of ornamental value of Lagerstroemia, more than 260 cultivars have been registered in the international authority of Lagerstroemia cultivars (http://www.usna.usda.gov/Research/Herbarium/Lagerstroemia/index.html). Due to the important value of Lagerstroemia, molecular tools have been employed to carry out identification of Lagerstroemia cultivars and interspecific hybrids (Pooler Citation2003; Pounders et al. Citation2007). Lagerstroemia macrocarpa (accession number: ZAFU 1507141) is deciduous tree with many-flowered terminal panicle which has valuable ornamental character and scattered in dry dipterocarp forest and open forests in Burma (Myanmar) and Laos (Khammouan). We got sample and specimens from Xishuangbanna Tropical Botanic Garden (XTBG) where L. macrocarpa has amount of introduction.
In contrast to large nuclear genomes, the chloroplast genome has a highly conserved circular DNA structure varying from 115 to 165 kb with uniparental inheritance, low recombination rates, conserved gene order and sequence similarity across the land plants (Palmer Citation1985; Ravi et al. Citation2008; Wicke et al. Citation2011). Plant chloroplast genomes have been a valuable source of informative markers in phylogenetic studies (Moore et al. Citation2010; Wang et al. Citation2010; Wu & Ge Citation2012), plant barcoding (Day & Goldschmidt-Clermont Citation2011) and biogeographical researches among populations (Wang et al. Citation2011). With the decreasing cost of next-generation sequencing approaches, sequencing whole chloroplast genomes is becoming more popular (Soltis et al. Citation2013). To date, more than 800 land plant species’ completed chloroplast genomes can be searched through the National Center for Biotechnology Information (NCBI) public database (Wu et al. Citation2015).
The raw Illumina reads were trimmed and filtered by quality score using Trimmomatic v0.3 (Bolger et al. Citation2014) with the following settings: leading: 3, trailing: 3, sliding window: 4:15 and minlen: 50. Then the CLC Genomics Workbench v7 (CLCbio) (http://www.clcbio.com) was employed to process de novo assembly of reads from L. macrocarpa with the default parameters. Three separate de novo assemblies (PE reads, single-end forward reads and single-end reverse reads) were made (Wu et al. Citation2015) and then combined into a single assembly. The whole chloroplast genome for L. macrocarpa was finished at 152,472 bp in length after combining the Sanger and Illumina sequence data. Through mapping the paired reads onto the finished genome, we verified our assembled length for the finished chloroplast genome with 1,473,293 (5% of the total reads) mapped reads across the whole chloroplast genome with at least 950 reads per position.
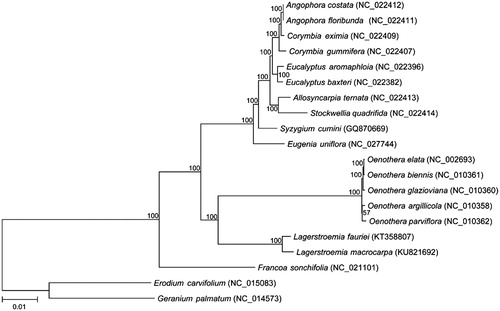
The final chloroplast genome was annotated with DOGMA (http://dogma.ccbb.utexas.edu/) and manually fixed exon-intron junctions (Wu et al. Citation2015). The complete chloroplast genome sequence was submitted to GenBank under the accession number of KU821692. Seventy shared protein coding genes were extracted and used to constructed the NJ tree with 500 bootstrap replicates by MEGA7 (www.megasoftware.net) () followed by the method used in (Wu et al. Citation2015).
Figure 1. The phylogenetic relationships among 18 species with two other species from Geraniaceae family as outgroup were constructed by neighbour-joining (NJ) method using MEGA7. The numbers on the branches are bootstrap values. GenBank accession numbers are listed following each species.
The L. macrocarpa chloroplast genome is 152,472 bp in size with 37.6% GC content, consists of a pair of inverted repeats of 17,562 bp, separated by a large single-copy region and a small single-copy region of 84,050 bp and 33,298 bp, respectively. 112 unique genes were annotated including 78 protein-coding genes, 4 ribosomal RNA genes and 30 transfer RNA genes. Among these protein-coding genes, eight genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, rps16), six tRNA genes with one intron each (trnAGUC, trnGUCC, trnIGAU, trnKUUU, trnLUAA, trnVUAC) and three genes (clpP, rps12 and ycf3) have two introns.
Funding information
This study was supported by The National Natural Science Foundation of China (No. 31300581).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Day A, Goldschmidt-Clermont M. 2011. The chloroplast transformation toolbox: selectable markers and marker removal. Plant Biotechnol J. 9:540–553.
- Egolf DR, Andrick AO. 1978. The Lagerstroemia handbook/checklist. Swarthmore (PA): American Association of Botanical Gardens and Arboreta.
- Furtado CX, Srisuko M. 1969. A revision of Lagerstroemia L. (Lythraceae). Gard Bull. 24:185–335.
- Koehne E. 1883. Botanische Jahrbücher für Systematik. Pflanzengeschichte Und Pflanzengeographie. 4:252–270.
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA. 107:4623–4628.
- Palmer JD. 1985. Comparative organization of chloroplast genomes. Annu Rev Genet. 19:325–354.
- Pooler MR. 2003. Molecular genetic diversity among 12 clones of Lagerstroemia fauriei revealed by AFLP and RAPD markers. HortScience. 38:256–259.
- Pounders C, Rinehart T, Sakhanokho H. 2007. Evaluation of interspecific hybrids between Lagerstroemia indica and L. speciosa. HortScience. 42:1317–1322.
- Qin H, Graham SA. 2007. Lagerstroemia. Flora China. 13:277–281.
- Ravi V, Khurana JP, Tyagi AK, Khurana P. 2008. An update on chloroplast genomes. Plant Syst E. 271:101–122.
- Soltis DE, Gitzendanner M, Stull G, Chester M, Chanderbali A, Jordon-Thaden I, Soltis PS, Schnable PS, Brad Barbazuk W. 2013. The potential of genomics in plant systematics. Taxon. 62:886–898.
- Wang L, Qi XP, Xiang QP, Heinrichs J, Schneider H, Zhang XC. 2010. Phylogeny of the paleotropical fern genus Lepisorus (Polypodiaceae, Polypodiopsida) inferred from four chloroplast genome regions. Mol Phylogenet E. 54:211–225.
- Wang L, Wu ZQ, Bystriakova N, Ansell SW, Xiang QP, Heinrichs J, Schneider H. 2011. Phylogeography of the Sino-Himalayan fern Lepisorus clathratus on “the roof of the world”. PLoS One. 6:e25896.
- Wang X, Wadl PA, Pounders C, Trigiano RN, Cabrera RI, Scheffler BE, Pooler MR, Rinehart TA. 2011. Evaluation of genetic diversity and pedigree within crapemyrtle cultivars using simple sequence repeat markers. J Amer Soc Hort Sci. 136:116–128.
- Wicke S, Schneeweiss GM, DePamphilis CW, Müller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76:273–297.
- Wu ZQ, Ge S. 2012. The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol Phylogenet Evol. 62:573–578.
- Wu ZQ, Tembrock LR, Ge S. 2015. Are differences in genomic data sets due to true biological variants or errors in genome assembly: an example from two chloroplast genomes. PLoS One. 10:e0118019.
