Abstract
In this study, complete chloroplast sequences of Artemisia gmelinii and Artemisia capillaris (the Asteraceae family), which have been used as herbal medicine in Korea, were characterized by de novo assembly with whole-genome sequence data. The genomes of A. gmelinii and A. capillaris were 151,318 bp and 151,056 bp in length, respectively. Both genomes harbored identical number of annotated genes, such as 80 protein coding genes, 4 rRNA genes and 30 tRNA genes. Phylogenetic tree revealed that both A. gmelinii and A. capillaris were closely grouped with other Artemisia species.
Main text
The genus Artemisia of the Asteraceae family includes over 500 species, of which A. gmelinii and A. capillaris, both called ‘Injin’ have long been used as a folk medicine in Korea (Abad et al. Citation2012; Wang et al. Citation2012). Previous studies have reported that A. gmelinii and A. capillaris contain useful metabolites with various pharmacological effects such as anti-infective and anti-oxidant activity and hepatoprotective and anti-fibrotic effects (Könczöl et al. Citation2012; Wang et al. Citation2012). However, to date, genetic and genome research related to barcoding system lack in Artemisia genus, despite the importance of herbal medicine authentication system is increased (Liu et al. Citation2013; Lee et al. Citation2016). In this study, we characterized complete chloroplast genome sequences of A. gmelinii and A. capillaris and analyzed phylogenetic relationship of the species, which will be useful for authentication and further molecular study in the Artemisia genus as well as in the Asteraceae family.
Complete chloroplast genome sequences of A. gmelinii and A. capillaris were established according to the previous reports (Kim et al. Citation2015a, Citationb). At first, we extracted DNA from leaves of A. gmelini and A. capillaris that were collected from Hongcheon-gun (Gangwon-Do, South Korea) and Taeanhaean National Park (the west coast of South Korea), respectively and maintained in Hantaek Botanical Garden (http://www.hantaek.co.kr/). Whole-genome sequencing was conducted using an Illumina HiSeq 2000 (Illumina, San Diego, CA). The high-quality paired end sequences of 2.1 and 1.6 Gb were generated from A. gmelinii and A. capillaris, respectively, and separately assembled using CLC genome assembler (v. beta 4.6, CLC Inc., Lyngby, Denmark). Afterwards, contigs representing chloroplast sequence were retrieved, ordered and combined into a single draft sequence by comparing chloroplast genomes of A. frigida (JX293720, Liu et al. Citation2013) and A. fukudo (KU360270, Lee et al. Citation2016). The complete genome of A. gmelinii and A. capillaris were deposited in GenBank (A. gmelinii: KU736962 and A. capillaris: KU736963).
Complete chloroplast genome sequences of A. gmelinii and A. capillaris were 151,318 bp and 151,056 bp in length, respectively. The genomes of the both species comprised of four parts such as a large single copy region, a small single copy region and pairs of inverted repeats, showed different length. A. gmelini was a large single copy region of 83,061 bp, a small single copy region of 18,335 bp and pairs of inverted repeats of 24,961 bp. A. capillaris was a large single copy region of 82,821 bp, small single copy region of 18,309 bp and pairs of inverted repeats of 24,963 bp. In both chloroplast genomes, identical number of genes were predicted through DOGMA annotation (http://dogma.ccbb.utexas.edu/) and BLAST searches, which consists of 80 protein coding genes, 4 rRNA genes and 30 tRNA genes.
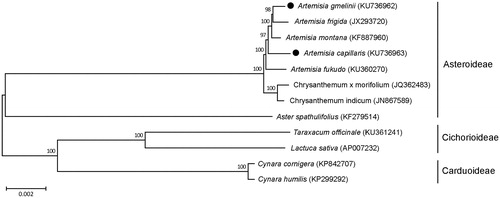
We further analyzed phylogenetic relationship of A. gmelinii and A. capillaris with other species belonging to the Asteraceae family (). Total 68 coding sequences were subjected to phylogenetic analysis using neighbour-joining (NJ) method in the MEGA 6.0 (Tamura et al. Citation2013). NJ phylogenetic tree exhibited that A. gmelinii and A. capillaris were grouped with other three Artemisia species, A. frigida, A. montana and A. fukudo, as expected ().
Figure 1. Phylogenetic tree showing relationship among A. gmelinii, A. capillaris and other 10 species belonging to the Asteraceae family. Total 68 coding region in chloroplast genome were used for analysis, and phylogenetic tree were generated using neighbour-joining method with 1000 bootstrap replicates in MEGA 6.0. The number above each node indicated bootstrap supporting values.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This work was supported by ‘15172MFDS246’ from Ministry of Food and Drug Safety in 2015, Republic of Korea.
References
- Abad MJ, Bedoyq LM, Apaza L, Bermejo P. 2012. The Artemisia L. genus: a review of bioactive essential oils. Molecules 17:2542–2566.
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ. 2015a. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One 10:e0117159.
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015b. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Könczöl Á, Béni Z, Sipos MM, Rill A, Háda V, Hohmann J, Máthé I, Balogh GT. 2012. Antioxidant activity-guided phytochemical investigation of Artemisia gmelinii Webb. ex Stechm.: isolation and spectroscopic challenges of 3,5-O-dicaffeoyl(epi)quinic acid and its ethyl ester. J Pharm Biomed Anal. 59:83–89.
- Lee YS, Park JY, Kim JK, Lee HO, Park HS, Lee SC, Kang JH, Lee TJ, Sung SH, Yang TJ. 2016. The complete chloroplast genome sequence of Artemisia fukudo Makino (Asteraceae). Mitochondrial DNA B Resour. Accepted.
- Liu Y, Huo N, Dong L, Wang Y, Zhang S, Young HA, Feng X, Gu YQ. 2013. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS One 8:e57533.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang JH, Choi MK, Shin JW, Hwang SY, Son CG. 2012. Antifibrotic effects of Artemisia capillaris and Artemisia iwayomogi in a carbon tetrachloride-induced chronic hepatic fibrosis animal model. J Ethnopharmacol. 140:179–185.
