Abstract
The Sikkim Mountain vole (Neodon sikimensis) belongs to the subfamily Arvicolinae. In this study, we sequenced the complete mitochondrial genome of N. sikimensis. It was determined to be 16,330 bp long and contained 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes and 1 control region. The nucleotide sequence data of 12 heavy-strand protein-coding genes of N. sikimensis and other 22 rodents were used for phylogenetic analysis. Tree constructed using Bayesian phylogenetic methods demonstrated that N. sikimensis was a sister to N. irene, and Microtus genus species did not cluster together with each other, making the genus a paraphyletic group.
The Rodentia has more than 2200 species in the word (Wilson & Reeder Citation2005). In China, there are more than 210 species (Jiang et al. Citation2015). The genus Neodon Horsfield 1841 is classified within the subfamily Arvicolinae (Musser & Carleton Citation2005), most of which lack comprehensive biological data. The Sikkim Mountain vole (Neodon sikimensis) belongs to the subfamily Arvicolinae, which is distributed in southern Tibet, Nepal, northeastern India and Bhutan (Smith et al. Citation2009). This species is restricted to alpine meadows and dense vegetation growing at the edges of rhododendron and coniferous forest, at the elevations of 2100–3700 m (Smith et al. Citation2009). It prefers to feed on green vegetation or seeds. In this study, we sequenced the complete mitochondrial genome of N. sikimensis (16, 330 bp; GenBank accession no. KU891252), to provide reference information for further study on Arvicolinae species.
This individual was captured in Motuo County, Tibet (latitude: 29°28′0.3″N, longitude: 94°59′59.3″E). A voucher specimen was deposited at the College of Life Sciences, Sichuan Normal University. The whole mitochondrial genome of N. sikimensis contains a set of 13 protein-coding genes, two ribosomal RNA genes (rRNA), 22 transfer RNA genes (tRNA) and 1 control region. The gene order and gene content of the mitochondrial genome of N. sikimensis is identical to that observed in most other Cricetidae species (Fan et al. Citation2011; Yang et al. Citation2012; Chen et al. Citation2015).
In order to explore the molecular phylogenetics evolution of Arvicolinae, the nucleotide sequence data of 12 heavy-strand protein-coding genes of N. sikimensis and other 22 rodents were used for the phylogenetic analysis. After alignment, the sequence set contained 10,788 bp. We used BEAST v1.7.4 (Drummond & Rambaut Citation2007) for Bayesian phylogenetic reconstructions for mitochondrial genomes of other 22 rodents. The best-fit GTR + I + G model of DNA substitution was obtained using jModelTest v2 (Darriba et al. Citation2012) under the Akaike Information Criterion (AIC). Sicista concolor was used as outgroup.
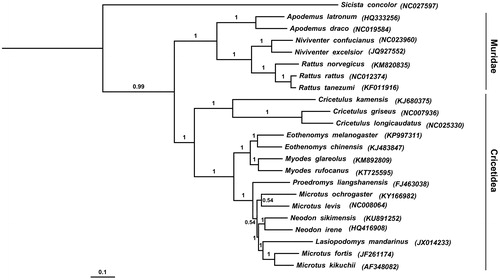
Tree constructed using Bayesian phylogenetic analysis is shown in . The 22 ingroup species sampled in this study belonged to two families: Muridae and Cricetidae. Neodon sikimensis clustered with species Neodon irene, Lasiopodomys mandarinus, Microtus fortis and Microtus kikuchii, formed a solid monophyletic group, well supported by a Bayesian posterior probability of 1.00 (). Bayesian analyses suggested that N. sikimensis was a sister to N. irene (Bayesian posterior probability =1.00). Furthermore, the BI tree demonstrated that Microtus levis, Microtus ochrogaster, Microtus fortis and Microtus kikuchii did not cluster together with each other. In order to have better understanding in the phylogenetic relationship within Arvicolinae, more complete mitochondrial genome sequences are required. We expect the study contributes to species identification and may facilitate further investigation of the molecular evolution of Arvicolinae.
Funding information
This research was funded by China Postdoctoral Science Foundation (2015M570801); The Key Project of Education Department of Sichuan Province (No. 15ZA0027); Experimental Technology and Management of Sichuan Normal University (No. SYJS2015-04); The Project of Chengdu Municipal Science and Technology Bureau (No. 2015-HM01-00134-SF).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Chen SD, Chen GY, Wei HX, Wang Q. 2015. Complete mitochondrial genome of the Père David’s Vole, Eothenomys melanogaster (Rodentia: Arvicolinae). Mitochondrial DNA. DOI: 10.3109/19401736.2015.1036246. [Epub ahead of print].
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. JModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 7:214–1471.
- Fan LQ, Fan ZX, Yue H, Zhang XY, Liu Y, Sun ZY, Liu SY, Yue BS. 2011. Complete mitochondrial genome sequence of the Chinese scrub vole (Neodon irene). Mitochondrial DNA. 22:50–52.
- Jiang ZG, Ma Y, Wu Y, Wang YX, Feng ZJ, Zhou KY, Liu SY, Luo ZH, Li CW. 2015. The checklist of China’s mammal species. Biodiversity. 3:351–364.
- Musser GG, Carleton MD. (2005). Family cricetidae. In Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. 3rd ed. Baltimore (MD): The Johns Hopkins Press. pp. 1189–1531.
- Smith A, Xie Y, Hoffmann RS, Lunde D, MacKinnon J, Wilson DE, Wozencraft WC, Gemma F. (2009). A guide to the mammals of china. Changsha (China): Hunan Education Publishing House.
- Wilson DE, Reeder DM. (2005). Mammal species of the world: a taxonomic and geographic reference. 3rd ed. Washington (DC): The Johns Hopkins University Press.
- Yang CZ, Hao HB, Liu SY, Liu Y, Yue BS, Zhang XY. 2012. Complete mitochondrial genome of the Chinese oriental vole Eothenomys chinensis (Rodentia: Arvicolinae). Mitochondrial DNA. 23:131–133.