Abstract
In this study, the complete mitochondrial genome of the smalleye whip ray Himantura microphthalma was first determined. The total length of the complete mitogenome is 17,636 bp, consisted of 37 genes with a typical gene order in vertebrate mitogenome. A total of 63 bp short intergenic spaces and 25 bp overlaps were found between 17 and 6 gene junctions. The overall base composition is 31.2% A, 29.2% T, 26.4% C, and 13.2% G. Two start codons (GTG and ATG) as well as two stop codons (TAG and TAA/T) were used in protein-coding genes. The phylogenetic result suggests that H. microphthalma should be assigned to the genus Dasyatis.
The smalleye whip ray Himantura microphthalma was first described briefly and assigned to the genus Dasyatis by Chen (Citation1948) according a single specimen collected from the Keelung market in Taiwan, which was assumed to be captured nearby China Sea. However, Compagno and Roberts (Citation1982) moved it to the genus Himantura based on its obscure ventral tail fold. Unfortunately, the holotype had been lost from the Taiwan Museum (Nishida & Nakaya Citation1988) and no further specimens have emerged. Therefore, the status of H. microphthalma has not been resolved (Manjaji-Matsumoto & Last Citation2006). In this study, we determined the complete mitochondrial genome and constructed the phylogenetic relationship of Dasyatidae based on mitogenomic data. We hope that it could be helpful to better understand the taxonomy and the phylogenetic position of H. microphthalma.
One specimen of H. microphthalma was collected from a pier in Dongshan island, Fujian province, China, which was considered to be captured from Taiwan Strait. The morphological diagnose of specimen was consistent with the description of Chen (Citation1948). The specimen was preserved in Xiamen University (voucher: DS2011031041) and the pictures were published (Liu et al. Citation2013). The experimental protocol and data analysis method were according to the previous study (Chen et al. Citation2013). Including H. microphthalma, 12 species of Myliobatiformes with the complete mitogenomes available were selected to construct the Bayesian tree.
The total length of the complete mitogenome of H. microphthalma was 17,636 bp (Genbank Accession Number: KF840390.1). The overall base composition of the H-strand was 31.2% A, 29.2% T, 26.4% C, and 13.2% G, with a relatively lower level of G and a slight AT bias of 60.4%. It has a typical mitogenomic organization and gene order similar to most vertebrates, containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 2 non-coding control region. There were totally 63 bp short intergenic spaces located in 17 gene junctions and 25 bp overlaps located in 6 gene junctions in the mitogenome. The 22 tRNA genes were interspersed along the genome, with the size ranging from 67 bp (tRNA-Ser2) to 75 bp (tRNA-Leu1). The sizes of 12S rRNA and 16S rRNA were 962 and 1700 bp, respectively. The 13 protein-coding genes use 2 start codons (GTG and ATG) as well as 2 stop codons (TAG and TAA/T), and most of them share common initial codon ATG and terminal codon TAA/T. The origin of light-strand replication (36 bp) was located between tRNA-Asn and tRNA-Cys genes. The control region was 1890 bp in length, rich in A + T (63.8%) and poor in G (14.1%).
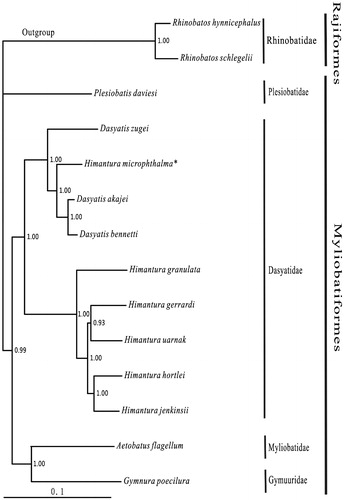
Most nodes of the Bayesian tree are strongly supported. Except for the Dasyatidae, which is monophyletic, the remaining three families within Myliobatiformes have one species available. However, H. microphthalma is inserted to the clade of Dasyatis instead of Himantura. In addition, the back and tail of H. microphthalma covered the smooth skin without denticles. Therefore, we support Chen’s classification that assigned this species to the genus Dasyatis.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding
This study was supported by the Special Scientific Research Funds for Central Non-profit Institutes, Chinese Academy of Fishery Sciences [2015B02XK02] and Guangdong Natural Science Foundation [2015A030310002].
Figure 1. Phylogenetic position of Himantura microphthalma. Rhinobatos hynnicephalus (NC_022841.1) and R. schlegelii (NC_023951.1) were selected as the outgroup. The 12 species of Myliobatiformes were: Plesiobatis daviesi (AY597334.1), Dasyatis zugei (NC_019643.1), D. akajei (NC_021132.1) and D. bennetti (KC633222.1), Himantura microphthalma* (KF840390.1), H. granulata (NC_023525.1), H. gerrardi (NC_026208.1), H. uarnak (NC_028325.1), H. hortlei (NC_027497.1), H. jenkinsii (KU873081), Aetobatus flagellum (NC_022837.1) and Gymnura poecilura (NC_024102.1).
References
- Chen JTF. 1948. Notes on the fish-fauna of Taiwan in the collections of the Taiwan Museum. I. Some records of platosomeae from Taiwan, with description of a new species of Dasyatis. Q J Taiwan Mus (Taipei). 1:1–14.
- Chen X, Ai W, Ye L, Wang X, Lin Yang S. 2013. The complete mitochondrial genome of the grey bamboo shark (Chiloscyllium griseum) (Orectolobiformes: Hemiscylliidae): genomic characterization and phylogenetic application. Acta Oceanol Sinica. 32:59–65.
- Compagno LJV, Roberts TR. 1982. Freshwater stingrays (Dasyatidae) of southeast Asia and New Guinea, with description of a new species of Himantura and reports of unidentified species. Environ Biol Fishes. 7:321–339.
- Liu M, Chen X, Yang S. 2013. Marine fishes of Southern Fujian, China. Beijing (China): Oceanpress. pp. 62.
- Manjaji-Matsumoto BM, Last PR. 2006. Himantura lobistoma, a new whipray (Rajiformes: Dasyatidae) from Borneo, with comments on the status of Dasyatis microphthalmus. Ichthyol Res. 53:290–297.
- Nishida K, Nakaya K. 1988. A new species of the genus Dasyatis (Elasmobranchii: Dasyatididae) from southern Japan and lectotype designation of D. Zugei Jpn J Ichthyol. 35:115–123.
