Abstract
In this study, the complete mitochondrial genome of Leiocassis argentivittatus has been determined by polymerase chain reaction methods for the first time. The overall base composition of Leiocassis argentivittatus mitogenome is 31.7% for A, 26.4% for C, 14.9% for G and 27.0% for T. The percentage of G + C content is 41.3%. The mitogenome is a circular DNA molecule of 16,534 bp in length with a D-loop region and contains 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes and 13 protein-coding genes. The mitochondrial genome sequencing for Leiocassis argentivittatus in this study provides important molecular data for further evolutionary analysis of Siluriformes.
Leiocassis argentivittatus, which belongs to order Siluriformes, family Bagridae, genus Leiocassis is a species of freshwater fish with body extention, slightly thick and compressed tail. It is mainly found in the Pearl River and Heilongjiang River system.
In this study, L. argentivittatus samples were collected from Fuyuan County, Heilongjiang Province of China (48°37′N; 134°28′E). The specimen is stored in Northeast Agricultural University and its accession number is PS71007001000100. The genome DNA was extracted following the traditional phenol–chloroform method (Taggart et al. Citation1992). Twenty-six primers were designed to amplify the PCR products for sequencing. The sequencing results were then assembled using ContigExpress 9.0 software (New York, NY). The transfer RNA (tRNA) genes were identified using the program tRNAscan-SE 1.21 (http://lowelab.ucsc.edu/tRNAscan-SE). The locations of protein-coding genes were determined by comparing with the corresponding known sequences of other Leiocassis fish species.
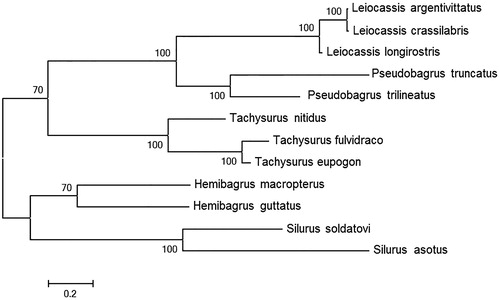
The complete mitochondrial genome length of L. argentivittatus was 16,534bp in length (GenBank accession number KX164404). It consisted of 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes and 1 D-loop region. The overall base composition of the mitogenome is 31.7% for A, 26.4% for C, 14.9% for G and 27.0% for T. The percentage of G + C content is 41.3%. To validate the phylogenetic position of L. argentivittatus, we performed multiple sequence alignment and MEGA 6.0 (Englewood, NJ) (Tamura et al. Citation2013) to construct a maximum-likehood tree containing complete mitochondrial genome DNA of 12 species in Siluriformes. As shown in the phylogenetic tree (), our sequence was clustered in genus Leiocassis, including Leiocassis crassilabris and Leiocassis longirostris.
Figure 1. A maximum-likelihood (ML) tree of the 12 species from Siluriformes was constructed based on complete mitochondrial genome data. The analyzed species and corresponding NCBI accession numbers are as follows: Leiocassis argentivittatus(KX164404), Leiocassis crassilabris (NC_021394), Leiocassis longirostris (NC_014586), Pseudobagrus truncatus (NC_021395), Pseudobagrus trilineatus (NC_022705), Tachysurus nitidus (NC_014859), Tachysurus fulvidraco (KC287172), Tachysurus eupogon (NC_018768), Hemibagrus macropterus (NC_019592), Hemibagrus guttatus (NC_023976), Silurus soldatovi (NC_014866), Silurus asotus (NC_015806).
Funding information
This work was supported by the National Natural Science under Grant number 31470131.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Taggart JB, Hynes RA, Prodoh PA, Ferguson A. 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol. 40:963–965.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
