Abstract
The nucleotide sequences of the two chloroplast (cp) genomes from Pogostemon stellatus and Pogostemon yatabeanus are the first to be completed in genus Pogostemon of family Lamiaceae. The structure of two Pogostemon cp genomes shows similar characteristic to the typical cp genome of angiosperms. The lengths of two cp genomes are 151,825bp and 152,707 bp, respectively. Two cp genomes are divided into LSC region (83,012 bp and 83,791 bp) and SSC region (17,524 bp and 17,568 bp) by two IR regions (25,644 bp and 25,674 bp). Both of two cp genomes contain 114 genes (80 protein-coding genes, 30 tRNA genes and 4 rRNA genes), 10 protein-coding genes and 7 tRNA genes duplicated in the IR regions. Similar to the typical cp genome of angiosperms, 18 of the genes in the Pogostemon cp genome have one or two introns. The overall A-T contents of two genomes are 61.8% which is also similar to general angiosperms. The A-T content in the non-coding (64.4%) is higher than in the coding (59.9%) regions. Sixty-seven and seventy-three simple sequence repeat (SSR) loci were identified in the P. stellatus and P. yatabeanus cp genomes, respectively. In phylogenetic analysis, genus Pogostemon shows closed relationship with Scutellaria baicalensis of Scutellarioideae
Results and discussion
Genus Pogostemon which included about 94 species is a large group of the Lamioideae clade in the family Lamiaceae (Panigrahi Citation1984; Chuakul Citation2002). None of the cp genome sequence have been published so far in genus Pogostemon, therefore we performed cp genome sequencing of Pogostemon stellatus (Lour.) Kuntze and Pogostemon yatabeanus (Makino) Press. Pogostemon stellatus and P. yatabeanus are aquatic perennial herbs which are highly acclaimed for ornamental plant in the aquarium trade. Specially, P. yatabeanus is one of the endangered species in Korea. The plant materials of P. stellatus and P. yatabeanus were collected from a single individual that lives in the natural wetland habitats in Jeju-island, South Korea. Voucher specimens and DNA samples were deposited in the Korea University Herbarium (KUS2014-1557, KUS2014-1822) and Plant DNA bank in Korea (PDBK 2014-1557, PDBK 2014-1822), respectively. Chloroplast genome sequences were analyzed using Illumina MiSeq (San Diego, CA), and assembled by Geneious 8.1.6 (Auckland, New Zealand) (http://www.geneious.com, Kearse et al. Citation2012). The complete cp genome sequences were submitted into NCBI database under the accession number of KP718620 for P. stellatus and KP718618 for P. yatabeanus, respectively. The lengths of complete cp genome sequence of P. stellatus and P. yatabeanus are 151,825bp and 152,707bp, respectively. The cp genome of P. stellatus is composed of 83,012bp of LSC region, 17,524bp of SSC region and 25,644bp of two IR regions, whereas the cp genome of P. yatabeanus is composed of 83,791bp of LSC region, 17,568bp of SSC region and 25,674bp of two IR regions. Both of two cp genomes consist of 114 individual genes which included 80 protein-coding genes, 30 transfer RNA genes and 4 ribosomal RNA genes. Among them, 10protein-coding genes and 7 tRNA genes are duplicated on the IR regions. Similar to the typical cp genome of angiosperms such as Panax and Sesamum, 18 of the genes in the each cp genome have one or two introns (Kim & Lee Citation2004; Yi & Kim Citation2012). Of these, rps12, clpP and ycf3 have two introns.
The major portion of the P. stellatus and P. yatabeanus cp genomes consist of gene-coding regions (59.2% and 58.9%) which consist of protein-coding region (52.8% and 52.6%) and RNA regions (6.4% and 6.3%), whereas the intergenic spacers (including 23 introns) comprise 40.8% and 41.1%, respectively. The overall A-T contents of two genomes are 61.8% which is similar to general angiosperms and other cp genomes of Lamiaceae (Shinozaki et al. Citation1986; Kim & Lee Citation2004; Cronn et al. Citation2008; Yi & Kim Citation2012; Zhu et al. Citation2014). In both of two genomes, the A-T content in the non-coding (64.4%) is higher than in the coding (59.9%) regions. The A-T contents of the IR region (56.6%) in two cp genomes are identical but the A-T contents in the LSC (63.7% and 63.8%) and SSC regions (67.9% and 68.0%) are slightly different. Sixty-seven and seventy-three SSR loci which were repeated more than 10 times identified in the P. stellatus and P. yatabeanus cp genomes, respectively.
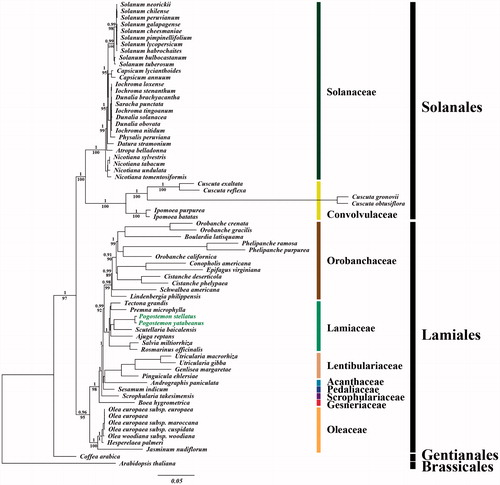
For the phylogenetic analysis, we assembled the 68 complete cp DNA sequences from the Solanales and Lamiales clade and two outgroup sequences from Gentianales and Brassicales clades. A total of 78 protein CDSs including rrn genes were aligned for the 70 analyzed taxa. The aligned data matrix consists of a total of 91,395bp. An ML tree was obtained with an − lnL = 578,964.0368 using the GTR + G + I base substitution model (). Similar to previous studies, genus Pogostemon forms a monophyletic group with Scutellaria baicalensis in Scutellarioideae and this monophyletic group shows close relationship with Ajuga reptans in Ajugoideae (Ingrouille & Bhatti Citation1998; Steane et al. Citation2004; Scheen et al. Citation2010; Bendiksby et al. Citation2011).
Figure 1. Chloroplast phylogenetic tree of the asterid I clade. A maximum-likelihood tree (−lnL= −578964.0368) inferred from analysis of alignment data containing 78 protein-coding genes in 70 chloroplast genome sequences by use of the GTR + Γ+I model. The numbers above and below each node indicate the Bayesian support percentages and bootstrap values, respectively. The scientific names with green color indicate two new sequences from P. stellatus (Lour.) Kuntze and P. yatabeanus (Makino) Press.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding information
This research was supported by the Eco-Innovation project (416-111-007) of the Ministry of Environment from KEITI to KJK.
References
- Bendiksby M, Thorbek L, Scheen AC, Lindqvist C, Ryding O. 2011. An updated phylogeny and classification of Lamiaceae subfamily Lamioideae. Taxon. 60:471–484.
- Chuakul W. 2002. A new combination in Pogostemon (Labiatae). Kew Bull. 57:1001–1003.
- Cronn R, Liston A, Parks M, Gernandt DS, Shen R, Mockler T. 2008. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 36:e122.
- Ingrouille M, Bhatti G. 1998. Infrageneric relationships within Pogostemon Desf.(Labiatae). Bot J Linn Soc. 128:159–183.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kim K-J, Lee H-L. 2004. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Research. 11:247–261.
- Panigrahi G. 1984. Nomenclatural notes on Pogostemon Desf. (Lamiaceae). Taxon. 33:102.
- Scheen AC, Bendiksby M, Ryding O, Mathiesen C, Albert VA, Lindqvist C. 2010. Molecular phylogenetics, character evolution, and suprageneric classification of Lamioideae (Lamiaceae). Ann Missouri Bot Gard. 97:191–217.
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, et al. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 5:2043–2049.
- Steane DA, de Kok RP, Olmstead RG. 2004. Phylogenetic relationships between Clerodendrum (Lamiaceae) and other Ajugoid genera inferred from nuclear and chloroplast DNA sequence data. Mol Phylogenet Evol. 32:39–45.
- Yi DK, Kim KJ. 2012. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS One. 7:e35872.
- Zhu A, Guo W, Jain K, Mower JP. 2014. Unprecedented heterogeneity in the synonymous substitution rate within a plant genome. Mol Biol Evol. 31:1228–1236.
