Abstract
The cone snail Conus textile belongs to the family Conidae. It is a kind of molluscivorous species. The complete mitochondrial DNA sequence was constructed by next-generation sequencing in this study. The mitogenome of C. textile is 15,765 bp in length, including 13 protein-coding genes, 22 tRNA genes, 2 ribosomal RNA genes and 1 control region. The base composition was 27.3% A, 37.9% T, 15.7% C and 19.1% G. The phylogenetic tree of C. textile with the other 6 Conus species and 15 Neogastropoda sea snails was built. It provides fundamental data for further research of phylogeny and biogeography with this genus.
The cone snails (Conus) are a species-rich genus of venomous marine gastropods. They inject their venom through a hypodermic needle-like radula harpoon that can penetrate deep into the dermis of their prey (Olivera, 1997). Their venoms consist of complex venoms composed mostly 100–250 disulfide-bridged peptides (Dobson et al. Citation2012). It’s a potent pharmacopoeia of individual bioactive peptide constituent, usually referred to as conotoxins or conopeptides (Bergeron et al. Citation2013). The cone snail Conus textile is a kind of molluscivorous sea snails (Röckel et al., 1995). We present the complete mitochondrial genome sequence of C. textile (Linnaeus Citation1758) in this study.
The specimens of C. textile (voucher no. 20141026-002; with Genbank accession no. KX155574) in this study were collected from north coast of Taiwan (25.203N, 121.695E). They are very common species in that area. The samples were deposited in Marine Toxins Lab., Department of Food Science, National Taiwan Ocean University, Taiwan. The total genomic DNA was extracted from muscle using magnetic bead technique with the KingFisher magnetic processors (ThermoFisher Scientific Inc., Worcester, MA). The raw next-generation sequencing reads generated from MiSeq sequencer (Illumina, San Diego, CA) were de novo assembled and reference mapping was conducted by commercial software (Geneious V9, Auckland, New Zealand) to produce a single circular form of complete mitogenome with about an average 24.3 coverage (1,340 out of 3,820,868 reads, 0.035%). The complete mitochondrial genome of C. textile is 15,765 bp in size, including 13 protein-coding genes, 22 tRNA genes, 2 ribosomal RNA genes (12S and 16S rRNA) and 1 control region. The overall base composition of C. textile is 27.3% for A, 37.9% for T, 15.7% for C and 19.1% for G. The protein coding rRNA and tRNA genes of C. textile mitogenome were predicted by using MITOS (Bernt et al. Citation2013) and tRNAscan-SE (Schattner et al. Citation2005).
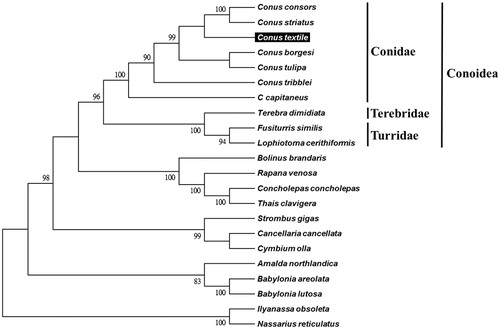
We used MEGA 6 (Tamura et al. Citation2013) to construct the phylogenetic relationships of the C. textile and related families by Neighbor-joining method with 1,000 bootstrap replicates based on the 13 protein-coding genes and 2 ribosomal RNA genes of the other 21 complete mitochondrial genomes of Neogastropoda sea snails, which are reported in Genbank of NCBI database. Bootstrap support values were relatively high, with 13 nodes having values >95% and 8 nodes demonstrating 100% bootstrap support (). C. textile was grouped together with other six Conus species from the family Conidae. The lineages of Conidae strongly supported in this report and agreed with previous studies (Bouchet et al. Citation2011; Puillandre et al. Citation2014).
Figure 1. Phylogenetic tree generated using the neighbor-joining method based on complete mitochondrial genomes. C. consors (KF887950), C. striatus (KX156937), C. textile (KX155574), C. borgesi (EU827198), C. tulipa (KR006970), C. tribblei (KT199301), C. capitaneus (KX155573), Terebra dimidiate (EU827196), Fusiturris similis (EU827197), Lophiotoma cerithiformis (DQ284754), Bolinus brandaris (EU827194), Rapana venosa (KM213962), Concholepas concholepas (JQ446041), Thais clavigera (DQ159954), Strombus gigas (KM245630), Cancellaria cancellata (EU827195), Cymbium olla (EU827199), Amalda northlandica (GU196685), Babylonia areolata (HQ416443), B. lutosa (KF897830), Ilyanassa obsoleta (DQ238598) and Nassarius reticulatus (EU827201).
Disclosure statement
Authors report no conflicts of interest. Authors alone are responsible for the content and writing of the paper.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bergeron ZL, Chun JB, Baker MR, Sandall DW, Peigneur S, Yu PY, Thapa P, Milisen JW, Tytgat J, Livett BG, et al. 2013. A ‘conovenomic’ analysis of the milked venom from themollusk-hunting cone snail Conus textile—The pharmacologicalimportance of post-translational modifications. Peptides. 49:145–158.
- Bouchet P, Kantor Y, Sysoev A, Puillandre N. 2011. A new operational classification of the Conidea (Gastropoda). J Molluscan Stud. 77:273–308.
- Dobson R, Collodoro M, Gilles N, Turtoi A, De Pauw D, Quinton L. 2012. Secretion and maturation of conotoxins in the venom ducts of Conus textile. Toxicon. 60:1370–1379.
- Linnaeus C. 1758. Systema Naturae per regna tria naturae, secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis. Editio decima, reformata. Holmiae: Impensis Direct. Laurentius Salvius. ii. p. 717.
- Olivera BM. 1997. E.E. Just Lecture, 1996. Conus venom peptides, receptor and ion channel targets, and drug design: 50 million years of neuropharmacology. Mol Biol Cell. 8:2101–2109.
- Puillandre N, Bouchet P, Duda TF, Jr., Kauferstein S, Kohn AJ, Olivera BM, Watkins M, Meter C. 2014. Molecular phylogeny and evolution of the cone snails. Mol Phylogenet E. 78:290–303.
- Röckel D, Korn W, Kohn AJ. 1995. Manual of the Living Conidae. Volume 1: Indo-Pacific Region. Germany: Verlag Christa Hemmen. p. 308–312.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:686–689.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
