Abstract
The complete mitochondrial genome of the silver chub M. storeriana was determined to be 16,709 bp and contained 22 tRNA genes, 2 rRNA genes and one control region. The whole genome base composition was 30.3% A, 28% T, 25.5% C and 16.2 G. This complete mitochondrial genome provides essential molecular markers for resolving phylogeny and aiding future conservation efforts.
Macrhybopsis chubs are a genus of the Leuciscinae subfamily consisting of small-bodied fishes that are typically obligate river species (Galat et al. Citation2005). The silver chub, M. storeriana, has been listed as Vulnerable (N3) in Canada due to anthropogenic disturbances such as eutrophication and baitfish harvesting (Boyko & Staton Citation2010). Conservation efforts have been complicated due to a wide variety of habitat utilization (Starrett Citation1950, Klutho Citation1983, Luttrell et al. Citation2002). The differences in habitat usage suggest that the taxonomy of the chubs may not have been fully resolved, which makes identifying populations that are susceptible to anthropogenic disturbances difficult.
Previous molecular studies have been unable to resolve the phylogeny of Macrhybopsis chubs with singular mitochondrial markers (Nagle & Simons Citation2012). Here, we report the complete mitogenome of the silver chub, M. storeriana. The silver chub was collected from a channelized section of the Missouri River along Nebraska’s eastern border (42°19'07.8”N 96°22'41.6”W) and is part of the ichthyology collection at the University of Kansas Biodiversity Institute (KUI 41379). This mitogenome will establish a solid basis to resolve phylogenetic confusion within this genus and may aid future conservation measures.
Genomic DNA was extracted and purified from fin tissue using the Qiagen DNeasy Blood and Tissue Kit (Hilden, Germany) for Genotyping by Sequencing (GBS). PCR free libraries were constructed with a TruSeq PCR Free library protocol and sequenced using an Illumina NextSeq500 (San Diego, CA) at the USGS Leetown Science Facility. Sequences were assembled using Velvet (Zerbino & Birney Citation2008), aligned with Mega 6.06 (Tamura et al. Citation2013) and annotated with MitoFish (Iwasaki et al. Citation2013). DOGMA was used to verify annotation and identify start and stop codons (Wyman et al. Citation2004).
The total length of the silver chub’s mitogenome was 16,709 bp (GenBank Accession No. KX139438). The mitogenome consisted of 22 tRNA genes, 2 rRNA genes and one control region. Fourteen of the tRNA genes were encoded on the heavy (H) strand along with all of the protein-coding genes except NADH dehydrogenase subunit 6. The whole genome base composition was 30.3% A, 28% T, 25.5% C and 16.2 G. The putative control region was located between tRNAPro and tRNAPhe and was 1056 bp long.
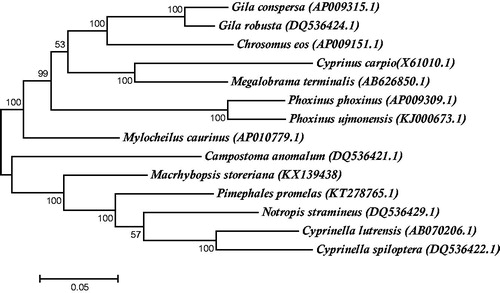
To investigate the position of M. storeriana within Leuciscinae, a maximum likelihood tree based on fourteen complete mitochondrial genomes was constructed using MEGA6 under the GTR + G + I model with 500 bootstrap replicates (Pattengale et al. Citation2010, Tamura et al. Citation2013) (). This maximum likelihood tree phylogenetically positioned M. storeriana as a sister clade to the Notropin clade supporting previous morphological phylogenetic analysis (Cavender & Coburn Citation1992).
Disclosure statement
The authors report no conflict of interest. The authors are solely responsible for the content and writing of this manuscript. Use of trade, product, or firm names does not imply endorsement by the U.S. Government.
Funding
This study was supported through funding from the University of Nebraska Omaha Graduate Research and Creative Activity (GRACA), UCRCA, Office of Graduate Studies Rhoden Fellowship and the Department of Biology.
References
- Boyko AL, Staton SK. 2010. Management plan for the Silver Chub, Macrhybopsis storeriana,in Canada. Species at risk act management plan series. Ottawa (CN): Fisheries and Oceans Canada, vi, p. 21.
- Cavender TM, Coburn M. 1992. Phylogenetic relationships of North American Cyprinidae. In: Mayden RL, editor. Systematics, historical ecology, and north american freshwater fishes. Stanford (CA): Stanford University Press. p. 293–327.
- Galat DL, Berry CR, Gardner WM, Hendrickson JC, Mestl GE, Power GJ, Stone C, Winston MR. 2005. Spatiotemporal patterns and changes in Missouri River fishes. Am Fish Soc Symp. 45:249–291.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, Nishida M. 2013. “MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Klutho MA. 1983. Seasonal, daily, and spatial variation of shoreline fishes in the Mississippi River at Grand Tower. Illinois: Southern Illinois University.
- Luttrell GR, Echelle AA, Fisher WL. 2002. Habitat correlates of the distribution of Macrhybopsis hyostoma (Teleostei: Cyprinidae) in western reaches of the Arkansas river basin. Trans Kansas Acad Sci. 105:153–161.
- Nagle B. C, Simons A. M. 2012. Rapid diversification in the North American minnow genus Nocomis. Mol Phylogenet Evol. 63:639–649.
- Pattengale N. D, Alipour M, Bininda-Emonds O. R, Moret B. M, Stamatakis A. 2010. How many bootstrap replicates are necessary? J Comput Biol. 17:337–354.
- Starrett W. 1950. Food relationships of the minnows and darters of the Des Moines River, Iowa. Ecology. 31:216–233.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. “MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman S. K, Jansen R. K, Boore J. L. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.