Abstract
The complete mitochondrial genome of Euthrix laeta (GenBank accession number KU870700) was sequenced by traditional PCR amplification and primer walking methods. The total length was 15,368 bp, including 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes and a A + T-rich region. The base composition of the genome was A (40.85%), T (39.34%), C (12.01%) and G (7.8%), respectively. The arrangement of all genes was identical to other lepidopteran insects. The phylogenetic relationships were established based on the nucleotide sequences of 13 protein-coding genes of mitochondrial genomes by the neighbor-joining method. The molecular-based phylogeny supported the traditional morphological classification on relationships within Lepidoptera species.
Euthrix laeta (Walker) (Lepidoptera: Lasiocampidae) is the wide pest which damaged bamboo and caused huger economic losses every year (Zhu et al. Citation2010). There have been many reports about integrated control of E. laeta in natural reserves (Chen & Chen Citation2008; Hao Citation2012). However, little study focused on its genetic characteristics. Therefore, it is very important to obtain the complete mitochondrial genome (mitogenome) sequence of E. Laeta for phylogenetic analysis of Lasiocampidae species.
The samples were collected from Taohong Ridge National Sika Deer Nature Reserve (116°42'10” E, 29°47'7” N), Jiujiang City, Jiangxi Province, China. The species identification was conducted by looking up the relevant literature and books (Liu & Wu Citation2006). The specimen was maintained in Natural Science Museum, College of Life Science, Nanchang University, Nanchang, China, with an accession number 150726NC. The adult of E. Laeta were removed wings and stored in 75% ethanol at −40 °C refrigerator for further use. Total genomic DNA was extracted from single specimen by using GENERAY BIOTECH Genomic DNA kit (Shanghai, China). The traditional PCR amplification and primer walking methods (Wei et al. Citation2009; Cao et al. Citation2012) were necessary. Six short fragments were amplified based on general primers of insects (Simon et al. Citation2006). According to these six specific fragments, we designed the primers of long fragments and amplified all the fragments. All the PCR products were sequenced by Sangon Biotech (Shanghai, China). Then, we obtained the complete mitogenome of E. Laeta. Genetic code, start and stop codons and codon bias were identified by being compared with the complete mitogenome of other Lasiocampidae species by Clustal X 1.83 software (Thompson et al. Citation1997) that had been submitted to NCBI and the 22 tRNA genes were identified by the online software MITOS Web Server (http://mitos.bioinf.uni-leipzig.de/index.py). In this study, the complete mitogenome of E. Laeta was first determined 15,368 bp in length with GenBank Accession NO. KU870700.
The complete mitogenome of E. laeta contained 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, 22 transfer RNA genes and a A + T-rich region. The arrangement of all genes was identical to other lepidopteran insects that had been regarded as a synapomorphy for Lepidoptera (Kim et al. Citation2010). ND5, ND4, ND4L, ND1 and eight tRNA genes (tRNAGln, tRNACys, tRNATyr, tRNAPhe, tRNAHis, tRNAPro, tRNALeu and tRNAVal) were encoded on the light strand; the remaining genes were encoded on the heavy strand. Except for COI with CGA start codon, respectively, the remaining PCGs initiated with the three orthodox start codons (ATN). The abnormal start codon in the COI gene was found presently in all mitochondrial genomes of Lepidoptera (Lee et al. Citation2006). Three PCGs (COI, COII and ND4) used incomplete stop codon T, which was commonly reported in other invertebrates (Masta & Boore Citation2004). The other 10 PCGs stopped with complete stop codon TAA. The length of 22 tRNA genes ranged from 64 to 71 bp. 16S rRNA was located between the tRNALeu and tRNAVal, 12S rRNA was located between tRNAVal and A + T-rich region on the light strand. The A + T-rich region was located between 12S rRNA and tRNAMet with a length of 372 bp. This region was believed to be involved in the regulation of transcription and control of DNA replication (Zhang & Hewitt Citation1997). The overall nucleotide composition of E. laeta was A (40.85%), T (39.34%), C (12.01%) and G (7.80%), which showed a high A + T bias.
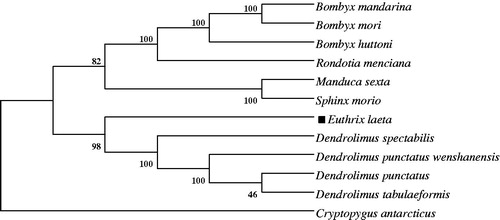
In addition, establishment of the phylogeny among Lepidoptera species was based on the nucleotide sequences of the 13 PCGs. As shown in , E. laeta was grouped with Dendrolimus punctatus wenshanensis (Lepidoptera: Lasiocampidae), Dendrolimus punctatus (Lepidoptera: Lasiocampidae), Dendrolimus spectabilis (Lepidoptera: Lasiocampidae) and Dendrolimus tabulaeformis (Lepidoptera: Lasiocampidae) in one branch; Manduca sexta and Sphinx morio were grouped in the family of Sphingidae; Bombyx mori, Bombyx huttoni, Rondotia menciana and Bombyx mandarina were grouped in the family of Bombycidae. Then the three branches were grouped with in the family of Lepidoptera, which was consistent with the morphological classification within the Lepidoptera insects.
Figure 1. Neighbor-Joining phylogeny based on the nucleotide sequences of 13 mitochondrial PCGs using MEGA 5.05 software (Tamura et al. Citation2011). The Collemola species, Cryptopygus antarcticus used as an outgroup. The genbank accession numbers of species used in phylogenetic tree, Bombyx mori (AC: AB083339); Bombyx mandarina (AC: AB070263); Manduca sexta (AC: NC_010266); Sphinx morio (AC: NC_020780); Bombyx huttoni (AC: NC_026518); Rondotia menciana (AC: KJ647172); Dendrolimus punctatus wenshanensis (AC: KJ913811); Dendrolimus punctatus (AC: NC_027156); Dendrolimus spectabilis (AC: KJ913815); Dendrolimus tabulaeformis (AC: NC_027157); Cryptopygus antarcticus (AC: NC_010533).
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This study was supported by National Natural Science Foundation of China (Grant number 31460553).
References
- Cao YQ, Ma C, Chen JY, Yang DR. 2012. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera. BMC Genomics. 13:276.
- Chen ZY, Chen ZM. 2008. A catalogue of insects from Dadongshan (Mt.) of Guangdong Nanlin national nature reserve (VI). J Environ Entomol. 2: 018.
- Hao NIU. 2012. Investigation of order lepidoptera insects in Wulingshan nature reserve. J Anhui Agri Sci. 4:049.
- Kim MJ, Wan X, Kim KG, Hwang JS, Kim I. 2010. Complete nucleotide sequence and organization of the mitogenome of endangered Eumenis autonoe (Lepidoptera: Nymphalidae). Afr J Biotechnol. 9:735–754.
- Lee E, Shin K, Kim M, Park H, Cho S, Kim C. 2006. The mitochondrial genome of the smaller tea tortrix Adoxophyes honmai (Lepidoptera: Tortricidae). Gene. 373:52–57.
- Liu YQ, Wu CS. 2006. Fauna Sinica: Insecta (Vol. 47) (Lepidoptera: Lasiocampidae). Beijing: Science Press, p. 336–339.
- Masta SE, Boore JL. 2004. The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol Biol Evol. 21:893–902.
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 37:545–579.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882.
- Wei SJ, Shi M, He JH, Sharkey MJ, Chen XX. 2009. The complete mitochondrial genome of Diadegma semiclausum (Hymenoptera: Ichneumonidae) indicates extensive independent evolutionary events. Genome. 52:308–319.
- Zhang DX, Hewitt GM. 1997. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol. 25:99–120.
- Zhu ZJ, Tu YH, Ye WX, Zhang AL, Bai HQ, Mao MH. 2010. Preliminary report on insect list damaged to bamboo in Huzhou. J Zhejiang Forest Sci Technol. 2:027.
