Abstract
The Chinese giant salamander, Andrias davidianus, is the biggest extant amphibian in the world. The population from Huangshan is distinct from other populations. The complete mitochondrial genome of a living wild-type Chinese giant salamander from Huangshan was sequenced. The total length is 16,565 bp, containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes and a D-loop. The phylogenetic tree of A. davidianus and 12 other closely species belonging to the order Caudata was reconstructed.
The family Cryptobranchidae has a combined Asian/North American distribution, but contains only three species (Andrias davidianus in China, A. japonicus in Japan, Cryptobranchus alleganiensis in North America). The Chinese giant salamander, A. davidianus, is the biggest extant amphibian in the world (Fei et al. Citation2006). A complete mitogenome of an individual (origin unknown) from Longsheng Chinese Giant Salamander Breeding Center in Guangxi province had been determined (Zhang et al. Citation2003).
The population from Huangshan is distinct from other populations, indicating the existence of localized divergence (Murphy et al. Citation2000). In the summer of 1994, a giant salamander was collected from Qimen County (29.75°N, 117.84°E), Anhui Province, China, and has been feeding in the Xiuning Institute of Rare Aquatic Animals (Voucher number: HS16091). During the time of experiment, its weight was 11.25 kg, total length was 1.20 m. In this article, the complete mitochondrial genome of this wild-type Chinese giant salamander has been determined and described in order to provide basic genetic information about this species.
The complete mitogenome of A. davidianus (Genbank accession number KX268733) was sequenced to be 16,565 bp, which consisted of 13 typical vertebrate protein-coding genes, 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes and 1 D-loop. The overall base composition of the entire genome was as follows: A (31.9%), T (32.7%), C (21.0%) and G (14.4%). All the 13 protein-coding genes initiated with ATG as the start codon, except for COI, which began with GTG. Nine protein-coding genes ended with complete stop codons (TAA, TAG, and AGG), and the other four genes terminated with T–– as an incomplete stop codon, which was presumably completed as TAA by post-transcriptional polyadenylation (Anderson et al. Citation1981). Among the mitochondrial protein-coding genes, the ATP8 was the shortest, while the ND5 was the longest. The 22 tRNA genes ranged in the size from 66 to 76 bp. The lengths of 12S and 16S rRNA are 925 bp and 1575 bp. The D-loop in size is 769 bp. As in most vertebrates, a non-coding spacer between the tRNA-Thr and tRNA-Pro genes is also found in A. davidianus (384 bp).
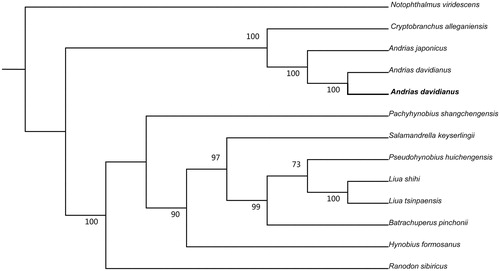
Mitochondrial genome sequences of A. davidianus in this study, and together with other 12 related species was used to perform the phylogenetic analysis. A maximum likelihood tree was constructed using online tool RAxML (Stamatakis et al. Citation2008) (). The phylogenetic analysis result was consistent with the previous research (Matsui et al. Citation2008).
Figure 1. A maximum likelihood (ML) tree of the 13 species from Caudata was constructed based on the dataset of mitochondrial genome by online tool RAxML. The numbers above the branch meant bootstrap value. Bold black branches highlighted the study species and corresponding phylogenetic classification. The analyzed species and corresponding NCBI accession number are as follows: Notophthalmus viridescens (EU880323), Cryptobranchus alleganiensis (GQ368662), Andrias japonicus (AB208679), Andrias davidianus (AJ492192), Andrias davidianus (KX268733), Pachyhynobius shangchengensis (DQ333812), Salamandrella keyserlingii (JX508762), Pseudohynobius huichengensis (FJ532060), Liua shihi (DQ333810), Liua tsinpaensis (KP233806), Batrachuperus pinchonii (KP122337), Hynobius formosanus (DQ333816), Ranodon sibiricus (AJ419960).
Acknowlegements
We thank Dr Hongguang Zha (Huangshan University) for data processing.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding information
This project was funded by the Natural Science Foundation of Anhui Province Education Department (No. KJ2014A244) and National Natural Science Foundation of China (No. 31471968).
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Insert Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290:457–465.
- Fei L, Hu SQ, Ye CY, Huang YZ. 2006. Fauna Sinica, Amphibia Vol. 1. General Accounts of Amphibia, Gymnophiona and Urodela. Beijing: Science Press.
- Matsui M, Tominaga A, Liu WZ, Tanaka-Ueno T. 2008. Reduced genetic variation in the Japanese giant salamander, Andrias japonicus (Amphibia: Caudata). Mol Phylogenetics Evol. 49:318–326.
- Murphy RW, Fu JZ, Upton DE, Lema T, De Zhao EM. 2000. Genetic variability among endangered Chinese giant salamanders, Andrias davidianus. Mol Ecol. 9:1539–1547.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web-servers. Systematic Biol. 75:758–771.
- Zhang P, Chen YQ, Liu YF, Zhou H, Qu LH. 2003. The complete mitochondrial genome of the Chinese giant salamander, Andrias davidianus (Amphibia: Caudata). Gene. 311:93–98.
