Abstract
The two complete mitochondrial genomes were sequenced from the monogonont rotifer Brachionus rotundiformis. The genome sequences were 10,268 bp and 11,703 bp in size, and the gene order and contents were identical with those of B. koreanus but were different in tRNA-Cys with B. plicatilis mitochondrial genomes. Of the 12 protein-coding genes (PCGs), five genes (ND1, ATP6, ND5, CO3, ND3) had incomplete stop codons. Furthermore, the start codon of ND4 and CO3 gene was ATA, while the start codon of other PCGs was ATG. The base composition of B. rotundiformis mitogenome shows an anti-G bias (12.05% and 10.24%) on the second and third position of the PCGs, respectively.
More than 30 species are described in the genus Brachionus (https://en.wikipedia.org/wiki/Brachionus). Of these, 15 species were deciphered for their phylogenetic placement through DNA taxonomy using CO1 and ITS1 genes in the Brachionus plicatilis species complex (Mills et al. Citation2016). However, to date, only a few complete mitochondrial genomes have been published in the genus Brachionus sp. (Suga et al. Citation2008 for B. plicatilis; Hwang et al. Citation2014 for B. koreanus). Brachionus rotundiformis (SS type) is a tropical euryhaline species (Hagiwara et al. Citation1995) and one of valuable food sources for rearing fish larvae with small mouth gape (Yúfera et al. Citation1997). The analysis of B. rotundiformis mitochondrial genome is important to identify laboratory stocks. In this paper, we report two complete mitochondrial genomes of the monogonont rotifer B. rotundiformis to better understand the phylogenetic placement and within the genus Brachionus.
The specimens were collected on Java Island, Indonesia in 1986 (kindly provided by Prof. Kazutsugu Hirayama, Nagasaki University, Japan) and maintained at the Laboratory of Professor Atsushi Hagiwara, Nagasaki University in Japan. The type was deposited in the ichthyological collection of the aculty of Fisheries, Nagasaki University (FFNU) under the accession no. FFNU-Rot-0001. We sequenced the whole genome of the rotifer B. rotundiformis from whole body genomic DNA of B. rotundiformis with paired-end libraries (300 bp, 500 bp, 800 bp) and mate-pair libraries (5 kb, 10 kb) using the Illumina HiSeq 2000 platform (GenomeAnalyzer, Illumina, San Diego, CA). De novo assembly was conducted by the ALLPATHS-LG release 44849 (http://www.broadinstitute.org/software/allpaths-lg/blog/). Sequenced reads with a phred score below 30 were removed. Of the assembled 2,390 B. rotundiformis scaffolds, two scaffolds were matched with the mitochondrial DNAs of B. koreanus (GenBank Nos. KC603851, KC603850). As a result, two mitochondrial genomes were obtained with full length.
The complete mitochondrial genomes of B. rotundiformis were 10,268 bp (mitochondrial DNA I; GenBank no. KX364936) and 11,703 bp (mitochondrial DNA II; GenBank no. KX364937). The direction of protein-coding genes (PGCs) was identical with those of B. koreanus of the genus Brachionus including the presence of nearly identical non-coding region (Identities: 3957/3964) (Suga et al. Citation2008; Hwang et al. Citation2013). Between the two species (B. rotundiformis, B. koreanus), the similarities of amino acids and nucleotides of 12 PCGs were 80.55% (96.12% for CO1 and 92.61% for Cytb) and 73.52% (82.21% for CO1 and 81.75% for Cytb), respectively. Of the 12 PCGs, five genes (ND1, ATP6, ND5, CO3, ND3) had incomplete stop codons as shown in B. plicatilis (Suga et al. Citation2008) and B. koreanus (Hwang et al. Citation2014). Particularly, in B. rotundiformis, there is an anti-G bias (12.05% and 10.24%) at the second and third position of codons. The start codon of ND4 and CO3 gene was ATA, while the start codon of other PGCs genes was ATG. The mitochondrial genome base composition of 12 PCGs was 24.45% for A, 45.40% for T, 13.77% for G and 16.38% for C. The A + T base composition (69.85%) was higher than G + C (43.65%).
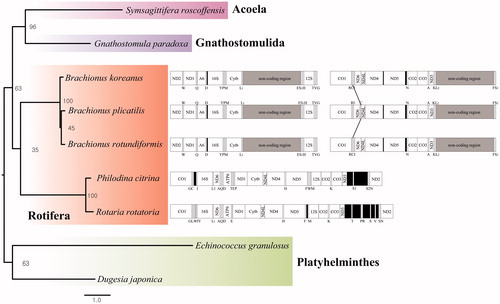
The placement of B. rotundiformis in the genus Brachionus with 11 mitochondrial DNA genes was shown in . B. rotundiformis clustered closely to B. plicatilis (L type) (Mills et al. Citation2016). Interestingly, in the genus Brachionus, tRNA-Cys was translocated between tRNA-Arg and tRNA-Ile in B. plicatilis, while tRNA-Cys of other Brachionus species was conserved in order in the mitochondrial genome. This indicates that the rearrangement of tRNAs is likely occurring in sporadic manner in the genus Brachionus.
Figure 1. Phylogenetic analysis. We conducted a comparison of the 11 mitochondrial DNA genes except for ND4L gene of Acoela, Gnathostomulida, Platyhelminthes, and Rotifera. The 11 mitochondrial DNA genes were aligned by ClustalW. Maximum likelihood (ML) analysis was performed by Raxml 8.2.8 (http://sco.h-its.org/exelixis/software.html) with GTR + Gamma + I nucleotide substitution model. The rapid bootstrap analysis was conducted with 10,000 replications with 48 threads running in parallel. The complete mitochondrial genomes were shown in parallel with a phylogenetic tree. The line on the mitochondrial genome indicates a translocation of tRNAs. The Platyhelminthes served as outgroup.
Disclosure statement
We thank Prof. Hans-Uwe Dahms for his comments on the manuscript. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Hagiwara A, Kotani T, Snell TW, Assava-Aree M, Hirayama K. 1995. Morphology, reproduction, genetics, and mating behavior of small, tropical marine Brachionus strains (Rotifera). J Exp Mar Biol Ecol. 194:25–37.
- Hwang D-S, Dahns H-U, Park HG, Lee J-S. 2013. A new intertidal Brachionus and intrageneric phylogenetic relationship among Brachionus as revealed by allometry and CO1-ITS1 gene analysis. Zool Stud. 52:13.
- Hwang D-S, Suga K, Sakakura Y, Park HG, Hagiwara A, Rhee J-S, Lee J-S. 2014. Complete mitochondrial genome of the monogonont rotifer, Brachionus koreanus (Rotifera, Brachionidae). Mitochondrial DNA. 25:29–30.
- Mills S, Alcántara-Rodríguez JA, Ciros-Pérez J, Gómez A, Hagiwara A, Hinson Galindo K, Jersabek CD, Malekzadeh-Viayeh R, Leasi F, Lee J-S, et al. 2016. Fifteen species in one: deciphering the Brachionus plicatilis species complex (Rotifera, Monogononta) through DNA taxonomy. Hydrobiologia. [Epub ahead of print]. doi:10. 1007/s10750-016-2725–7
- Suga K, Mark Welch DB, Tanaka Y, Sakakura Y, Hagiwara A. 2008. Two circular chromosomes of unequal copy number make up the mitochondrial genome of the rotifer Brachionus plicatilis. Mol Biol Evol. 25:1129–1137.
- Yúfera M, Parra G, Pascual E. 1997. Energy content of rotifers (Brachionus plicatilis and Brachionus rotundiformis) in relation to temperature. Hydrobiologia. 358:83–87.
