Abstract
The complete mitochondrial genome of the European blackfly, Simulium variegatum Meigen, 1818 was sequenced using a combined Illumina and Sanger sequencing approach. Using the known sequence of Chironomus tepperi Skuse, 1889 (Chironomidae) homologous NGS reads were identified and assembled. The genome is 15,367 bp in length and includes 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes and a control region. Gene order resembles that of the ancestral dipteran gene arrangement. The base composition of the genome is A (37.6%), T (35.3%), C (15.8%) and G (11.3%). The control region between 12S rRNA and tRNAIle is composed of 362 bp with no obvious repetitive motifs.
Blackflies belong to the family Simuliidae, a group of highly specialized aquatic insects. Adult females search for a blood meal and are notorious vectors of blood borne disease, most notably river blindness, onchocerciasis. Simulium variegatum (Meigen, 1818) is commonly found in small streams to large rivers throughout Ireland and the north and west of Britain. S. variegatum takes blood from dogs and livestock as well as man (Davies & Williams Citation1962). Their range extends from Algeria and Portugal in the West, throughout Europe through to Iran and the Caucasus in the East and with fourteen synonyms may be composed of a species complex (Adler & Crosskey Citation2015).
The S. variegatum specimen used in this study was collected from the River Alyn at Llandegla, Denbighshire (53.063214, -3.201966) on the 25 June 2014 as a pupa and preserved in absolute ethanol. DNA was sequenced on a HiSeq 2500 sequencer (Illumina) (San Diego, CA) as part of an environmental genomics project. The pupal cocoon was archived at CEH under the voucher code CEHSIMJD0822. The genome was assembled with Geneious 8.1.7 (Auckland, New Zealand) (Kearse et al. Citation2012) using the mitogenome of Chironomus tepperi (Acc. No. JN861749) as a reference. The first draft genome was used as a search query to determine additional reads in the 10 million read database. Finally, PCR and Sanger sequencing was used to validate the accuracy of ambiguous regions located in the non-coding region. A combination of primers (available from the author) was developed to amplify the entire control region of the S. variegatum mitochondrial genome. The mitogenome was annotated for CDS, 16S, 18S and the control region by comparison with the C. tepperi genome and using the ORF finder function in Geneious and the MITOS web server (Bernt et al. Citation2013). For tRNA prediction, the programs ARWEN v1.2.3 and tRNAscan-SE 1.21 were used (Lowe & Eddy Citation1997; Laslett & Canback Citation2008). The complete mitogenome of S. variegatum was assembled using a combination of Illumina Hiseq (San Diego, CA) and Sanger sequences. The final assembly contains 9593 Illumina sequences, 0.2% of the total dataset with an average sequence coverage of 113. The mitochondrial genome includes 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes and a control region between 12S rRNA and tRNAIle, composed of 361 bp containing no apparent repetitive units. The base composition of the genome was determined to be A (37.6%), T (35.3%), C (15.8%) and G (11.3%) with a GC content of 27.0%. The annotated mitogenome, with a length of 15,367 bp, is available online at NCBI (GenBank accession number KU252587) and shares the highest sequence identity to the mitochondrial genome of Simulium aureohirtum ().
Figure 1. A maximum likelihood phylogenetic tree of the mitochondrial genomes Simulium variegatum (shown boxed) and forty six further Nematocera taxon sequences retrieved from GenBank. Sequence alignments were conducted using ClustalW and the tree constructed using PhyML with 100 bootstrap replicates using the GTR + I + G model of substitution. An alignment of 14114 characters was used for the analysis incorporating the following sequences: Aedes aegypti EU352212; Aedes albopictus AY072044; Aedes notoscriptus KM676218; Anopheles albitarsis HQ335344; Anopheles arabiensis KT382816; Anopheles atroparvus KT382817; Anopheles bellator KU551287; Anopheles christyi KT382818; Anopheles coluzzii KT382819; Anopheles cracens JX219733; Anopheles cruzii KJ701506; Anopheles culicifacies KT382820; Anopheles darlingi GQ918272; Anopheles deaneorum HQ335347; Anopheles epiroticus KT382821; Anopheles gambiae str. PEST G3 L20934; Anopheles hinesorum JX219734; Anopheles homunculus KU551283; Anopheles laneanus KU551288; Anopheles maculatus KT382822; Anopheles melas KT382823; Anopheles merus KT382824; Anopheles minimus KT382825; Anopheles punctulatus KT382826; Anopheles quadrimaculatus L04272; Anopheles sinensis KT218684; Anopheles stephensi KT382827; Arachnocampa flava JN861748; Chironomus tepperi JN861749; Cramptonomyia spenceri JN861747; Culex pipiens pipiens HQ724614; Culex quinquefasciatus GU188856; Culex tritaeniorhynchus KT852976; Dixella aestivalis KT878382; Haemagogus janthinomys KT372555; Nyssomyia umbratilis KP702938; Ochlerotatus vigilax KP721463; Phlebotomus chinensis KR349297; Phlebotomus papatasi KR349298; Polypedilum vanderplanki KT251040; Protoplasa fitchii JN861746; Ptychoptera species JN861744; Rhopalomyia pomum GQ387649; Simulium aureohirtum KP793690; Sylvicola fenestralis JN861752; Trichocera bimacula JN861750.
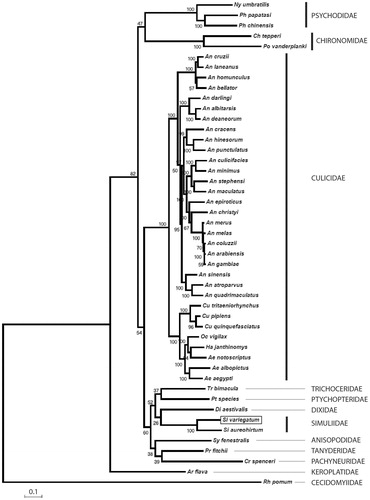
We provide the first complete mitochondrial genome from a European simuliid. Little is known about the molecular biology of blackflies in comparison with other medically important Diptera. A recent study illustrated the paucity of blackfly sequences available in GenBank in comparison to other Diptera (Adler et al. Citation2010). However, a blackfly BAC library is now available (Crainey et al. Citation2010) and a current blackfly genome project is in development (Brockhouse Citation2015). The availability S. variegatum mitogenome will provide a useful resource for understanding Simuliidae evolution.
Acknowledgements
We would like to thank Scot E. Dowd for providing the Illumina sequencing dataset.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding
This study was supported by a NERC innovation A grant.
References
- Adler PH, Cheke RA, Post RJ. 2010. Evolution, epidemiology, and population genetics of black flies (Diptera; Simuliidae). Infect Genet Evol. 10:846–865.
- Adler PH, Crosskey RW. 2015. World blackflies (Diptera: Simuliidae): a comprehensive revision of the taxonomic and geographical inventory. 123 pp. Available from: http://www.clemson.edu/cafls/biomia/pdfs/blackflyinventory.pdf (accessed 8 November 2015).
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Brockhouse C. 2015. An update on the Simulium genome project. British Simuliid Group Bulletin. 44:8–9.
- Crainey JL, Hurst J, Wilson MD, Hall A, Post RJ. 2010. Construction and characterisation of a BAC library made from field specimens of the onchocerciasis vector Simulium squamosum (Diptera: Simuliidae). Genomics. 96:251–257.
- Davies L, Williams CB. 1962. Studies on black flies (Diptera: Simuliidae) taken in a light trap in Scotland. Trans R Ent Soc London. 114:1–47.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Meigen JW. 1818. Systematische Beschreibung der Bekannten Europäischen Zweiflügeligen Insekten Vol. 1. Forstmann, Aachen, 332 pp.
