Abstract
The nemacheilid genus Triplophysa is widely distributed in the Qinghai-Tibet Plateau and its adjacent areas. The complete mitochondrial genome of the loach Triplophysa stenura collected from the Peigucuo Lake was sequenced. The genome was 16,569 bp in length, including 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and a non-coding control region (D-loop). Most of the genes were encoded on the heavy strand, except for ND6 and eight tRNA genes. The overall base composition of T. stenura was 27.8% for A, 28.4% for T, 25.4% for C, 18.4% for G, with a slight A + T rich feature (56.2%). Phylogenetic analyses showed that all Triplophysa species clustered together and T. stenura formed a sister relationship with T. tibetana. The complete mitogenome data of T. stenura would provide essential information in understanding phylogenetic relationships among Triplophysa species.
The genus Triplophysa is a strongly diverged fish group in the family Nemacheilidae, with 140 valid species, more than 80% of which are known from China so far (Ren et al. Citation2012; Froese & Pauly Citation2016). However, less than 20 complete mitogenome sequences of this genus are available from GenBank until now. Triplophysa stenura occurs in the Qinghai-Tibet Plateau, upper Yangtze, Mekong, Salween and Brahmaputra drainages (Kottelat Citation2012; Zhang et al. Citation2015). Here we determined the complete mitochondrial genome sequence of T. stenura (16569bp; GenBank accession number KX354975) and analyzed its phylogenetic position within genus Triplophysa.
Samples of T. stenura were collected from Peigucuo Lake (29°1′N, 85°34′13″E), a tributary of the Yarlung Zangbo River in Tibet, and preserved in the Key Laboratory of Freshwater Fish Reproduction and Development (Southwest University, China). We designed eight sets of primers and methods for sequence analyses strategies followed the procedures outlined in Wang et al. (Citation2012).
The complete mitogenome of T. stenura was a circular molecule with 16,569 nucleotides, including 13 protein-coding genes, 2 ribosomal RNA genes, 22 tRNA genes, and a non-coding control region (D-loop), which demonstrated a typical vertebrate mitogenome feature (Peng et al. Citation2006). Most of the genes were encoded on the heavy strand, except for ND6 and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAGlu and tRNAPro). The rank of overall nucleotide composition was T (28.4%)>A (27.8%)>C (25.4%)> G (18.4%), and A + T content (56.17%) was higher than G + C content (43.83%). All of the 13 protein-coding genes showed the regular initiation codon ATG with the sole exception of COI (started with GTG). Furthermore, the stop codons for five of the 13 protein-coding genes were TAA (ND1, COI, ATP6, ATP8, ND4L) and two of them were TAG (ND5, ND6), whereas other six genes possessed incomplete stop codons TA or T (ND2, ND3, ND4, COII, COIII and cytb). The gene arrangements were similar with some other Triplophysa species (e.g. Lei et al. Citation2016).
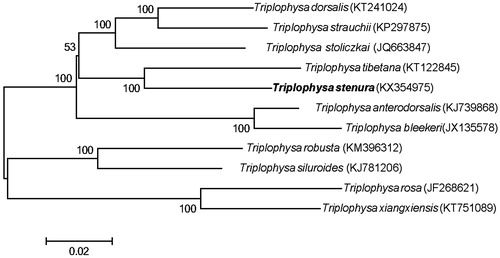
In order to explore the phylogenetic position of T. stenura, we retrieved other available mitogenome sequences of Triplophysa species and constructed a neighbour-joining tree in MEGA 6.0 (Tamura et al. Citation2013). The result strongly supported that T. stenura formed a sister relationship with T. tibetana (). Therefore, we expect that our new mitogenome data of T. stenura will contribute to elucidate the phylogenetic relationships among all the species of Triplophysa.
Acknowledgements
We thank Dr. Kai Zhao for the sample collection.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding
This work was supported by the Fund of Key Laboratory of Freshwater Fish Reproduction and Development [Ministry of Education; Grant no. FFRD-2010-01].
References
- Froese R, Pauly D. 2016. FishBase. World Wide Web electronic Publication. version (06/2016). Available from: http://www.fishbase.org.
- Kottelat M. 2012. Conspectus cobitidum: an inventory of the loaches of the world (Teleostei: Cypriniformes: Cobitoidei). Raffles Bull Zool. 26:1–199.
- Lei DJ, Kanu UC, Zhao G, Xie P, Yuan H, Niu JG, Ma XF. 2016. The complete mtDNA genome of Triplophysa dorsalis (Cypriniformes, Balitoridae, Cobitoidea): genome characterization and phylogenetic analysis. Mitochondrial DNA. 27:3745–3746.
- Peng ZG, Wang J, He SP. 2006. The complete mitochondrail genome of the helmet catfish Cranoglanis bouderius (Siluriformes: Cranoglanididae) and the phylogeny of otophysan fishes. Gene. 376:290–297.
- Ren Q, Yang JX, Chen XY. 2012. A new species of the genus Triplophysa (Cypriniformes: Nemacheilidae), Triplophysa longliensis sp.nov, from Guizhou, China. Zootaxa. 3586:187–194.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. Mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang JJ, Tang QY, Wang ZJ, Zhang YG, Wu Q, Peng ZG. 2012. The complete mitogenome sequence of a cave loach Triplophysa rosa (Teleoste, Balitoridae, Nemacheilinae). Mitochondrial DNA. 23:366–368.
- Zhang CG, Zhao YH, Xing YC, Zhou W, Tang WQ. 2015. Species diversity and distribution of inland fishes in China. Beijing: Science Press. (in Chinese).