Abstract
The complete mitochondrial genome (mitogenome) sequence of Eophona migratoria was determined by shotgun sequencing and reported for the first time using the muscle tissue. The total length of the sequence is 16,798 bp, which contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, 1 control region, and 1 extra pseudo-control region. A phylogenetic analysis on the basis of 12 protein-coding genes of 14 passeriformes species’ mitochondrial genomes using maximum-likelihood (ML) demonstrated that Coccothraustes coccothraustes is the most close to E. migratoria.
The Eophona migratoria is a kind of granivorous passerine in the family Fringillidae, which is mainly distributed in the eastern Siberia, eastern China, Korean Peninsula, and southern Japan (Zuccon et al. Citation2012). Fringillidae is the most abundant avian family, which account for approximately 20% of total passerine diversity and 10% of the total avian diversity (Yuri & Mindell Citation2002). In this study, the complete mitochondrial genome sequence of E. migratoria was sequenced by Chain Termination Method and reported for the first time using muscle tissue. The specimen of E. migratoria was collected from Zhumadian City, Henan Province, China (32.98°N, 114.02°E). The sample died due to poaching activities and was stored at laboratorial ultra-low temperature freezer in 2015. All sampling procedures and experimental manipulations held the proper permits.
The complete mitogenome of E. migratoria (Genbank accession number KX423959) was sequenced to be 16,798 bp. Sequence analysis showed that the genome structure is consistent with other Passeriformes species, which contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, 1 control region, and 1 extra pseudocontrol region. The structure of the mitogenome is a typical vertebrate mitochondrial gene arrangement (Boore Citation1999; Zhang et al. Citation2016; Zhao et al. Citation2016). Nine genes were encoded on the L-strand, including ND6 and 8 tRNA genes, and the remaining 28 genes were encoded on the H-strand. The position of the genes were predicted and identified by comparing with the corresponding genes of Coccothraustes coccothraustes, which is considered as the closely related species (Zuccon et al. Citation2012). Sequence analysis showed its total base composition as follows: C (34.5%), A (29.4%), T (23.5%), and G (12.6%); the percentage of A and T (52.9%) is higher than G and C (47.1%), which is consistent with other vertebrate mitogenome (Yang et al. Citation2015).
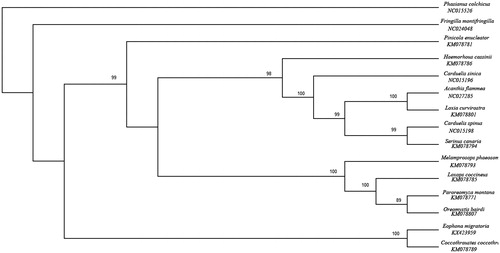
Phylogenetic relationships of E. migratoria and 13 other Passeriformes species were analyzed using the maximum-likelihood (ML) method based on the 12 protein-coding genes (except ND6 gene), with Phasianus coichicus (NC015526) used as an outgroup. GTR + I + G was chosen as the optimal evolutionary model according to the AIC test criterion by MrModeltest 3.7 (Tracey et al. Citation2007). A maximum-likelihood (ML) tree was constructed based on the dataset by PAUP 4.0, and the parameter of the bootstrap was determined as 100 replicates (Felsenstein Citation1981; Wilgenbusch & Swofford Citation2003; Zhang et al. Citation2016). The result of the phylogenetic tree showed that Fringilla montifringilla belonging to genus Fringilla is on the base in this tree (), and the position of the birds are mostly identical with the phylogenetic analyses based on Cytb gene (Marshall & Baker Citation1998). In this tree, E. migratoria is clustered with C. coccothraustes, which indicate that they have a much closer relationship (Questiau et al. Citation1999; Zuccon et al. Citation2012). The phylogenetic tree reveals that the data of our new determined mitogenome explain some related evolutionary issues. We expect this study to provide a useful database for further research in Fringillidae.
Figure 1. ML tree of 14 species from Fringillidae was constructed based on the dataset of 12 mitochondrial protein-coding genes (except ND6 gene). Numbers on top of branches meant bootstrap value. Accession numbers for the published sequences in GenBank are as follows: E. migratoria (KX423959), F. montifringilla (NC024048), C. coccothraustes (KM078789), Acanthis flammea (NC027285), Loxia curvirostra (KM078801), Carduelis spinus (NC015198), Serinus canaria (KM078794), C. sinica (NC015196), Haemorhous cassinii (KM078786), Paroreomyza Montana (KM078771), Oreomystis bairdi (KM078807), Loxops coccineus (KM078785), Melamprosops phaeosoma (KM078793), Pinicola enucleator (KM078781), Phasianus colchicus (NC015526).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 17:368–376.
- Marshall HD, Baker AJ. 1998. Rates and patterns of mitochondrial DNA sequence evolution in fringilline finches (fringilla spp.) and the greenfinch (Carduelis chloris). Mol Bio Evol. 15:638–646.
- Questiau S, Gielly L, Clouet M, Taberlet P. 1999. Phylogeographical evidence of gene flow among common crossbill (Loxia curvirostra, Aves, Fringillidae) populations at the continental level. Heredity. 83:196–205.
- Tracey SR, Smolenski A, Lyle JM. 2007. Genetic structuring of Latris lineata at localized and transoceanic scales. Mar Biol. 152:119–128.
- Wilgenbusch JC, Swofford D. 2003. Inferring evolutionary trees with PAUP. Curr Protoc Bioinformatics. Chapter 6:Unit 6.4. doi: 10.1002/0471250953.bi0604s00.
- Yang DC, Sun Y, Lu CH. 2015. The complete mitochondrial genome of Zosterops japonicas (Aves, Zosteropidae). Mitochondrial DNA. [Epub ahead of print]. doi: 10.3109/19401736.2015.1101581.
- Yuri T, Mindell DP. 2002. Molecular phylogenetic analysis of Fringillidae, “New World nine-primaried oscines” (Aves: Passeriformes). Mol Phylogenet Evol. 23:229–243.
- Zhang H, Dou H, Yang X, Zhao C, Liu G, Zhang J. 2016. The complete mitochondrial genome sequence of the Sparrow hawk (Accipiter nisus). Mitochondrial DNA Part A. 27:1648–1649.
- Zhao C, Zhang H, Liu G, Yang X, Zhang J. 2016. The complete mitochondrial genome of the Tibetan fox (Vulpes ferrilata) and implications for the phylogeny of Canidae. Comptes Rendus Biologies. 339:68–77.
- Zuccon D, Prŷs-Jones R, Rasmussen PC, Ericson PG. 2012. The phylogenetic relationships and generic limits of finches (Fringillidae). Mol Phylogenet Evol. 62:581–596.
