Abstract
The complete mitochondrial genome of Pycnarmon lactiferalis (Walker, 1859) has been determined. The entire sequence is 15,219 bp in length which contains 13 protein-coding genes (PCGs), 22 transfer RNA genes, two ribosomal RNA genes, and an A + T-rich region. All PCGs start with the typical ATN codon except for COI with CGA. TAA is used for all the PCGs, while the COI, COII and ND5 possess incomplete termination codons T or TA. The secondary structure of 22 tRNAs have the typical clover-leaf pattern except for tRNASer(AGN) lacking the dihydrouridine (DHU) stem. The A + T-rich region, located between the srRNA and tRNAMet, do not contain the motif “ATAGA” that is conserved across other lepidopteran species mitogenomes. A phylogenetic relationship of Pyraloidea has been reconstructed based on 13 PCGs data using Bayesian inference (BI) method.
Pycnarmon lactiferalis (Walker, 1859) is a species of the family Crambidae which contains various agriculture and forest pests. At present, only 23 complete mitochondrial genome sequences are available from Crambidae in GenBank database. The complete mitochondrial genome sequence of P. lactiferalis had been determined in this study. The new date of the sequence could provide useful information for population genetics and evolutionary studies.
Specimens of P. lactiferalis were collected from Xunyangba (33.33°N; 108.33°E), Ningshan County, Shaanxi province in the Qinling Mountain region of central China. DNA was extracted from the adult P. lactiferalis. Voucher specimens have been deposited in the Insect Collection (Accession Number SNU-Lep-20150021-5), College of Life Sciences, Shaanxi Normal University, Xi’an, China 710062.
The complete mitochondrial genome of P. lactiferalis is 15,219 bp in size with 81.68% A + T content. It consists of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes and an A + T-rich region. Gene order is identical with other Pyraloidea species (Chai et al. Citation2012; Ye et al. Citation2013; Cao & Du Citation2014). There are 14 intergenic regions (except for the A + T-rich region) ranging from 1 to 47 bp in P. lactiferalis mitochondrial genome. The longest intergenic spacer is located between the tRNAGln and ND2, with an extremely high AT content. Another intergenic spacer, located between the tRNASer(UCN) and ND1, contained the motif “ATACTAA” that is conserved across other lepidopteran species mitochondrial genomes (Cameron & Whiting Citation2008; He et al. Citation2015).
All PCGs begin with ATN codon, except for COI, which starts with the CGA. COI and COII genes end with T and the ND5 gene with TA as incomplete stop codon, and the remaining genes stop with TAA. The 22 tRNA genes are ranged in size from 62 to 71 bp. The large ribosomal subunit (lrRNA) and small ribosomal subunit (srRNA) genes are 1343 and 777 bp in size, located between tRNALeu(CUN) and AT-rich region, separated by tRNAVal gene. The length of the AT-rich region is 339 bp with 94.69% A + T content, which is between srRNA and tRNAMet.
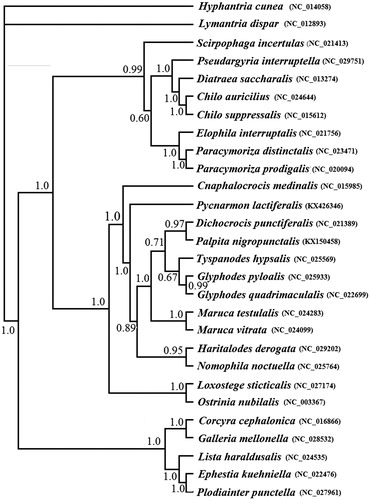
A phylogenetic analysis has been performed based on the 13 PCGs dataset using BI analysis (). A monophyly of the families Pyralidae and Crambidae is well supported. Within Crambidae, P. lactiferalis and other Spilomelinae species were clustered together, and this branch clustered with Pyraustinae species. The clear evolutionary relationships of them were required more taxon samples from each subfamily.
Nucleotide sequence accession number
The complete mitochondrial genome sequence of P. lactiferalis has been assigned with GenBank accession number KX426346.
Acknowledgements
The authors would like to thank M.S. Fei Ye (College of Life Sciences, Shaanxi Normal University) for assistance with sequencing and annotating. This research was supported in part by the National Natural Science Foundation of China (31372158).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
References
- Cameron SL, Whiting MF. 2008. The complete mitochondrial genome of the tobacco hornworm, Manduca sexta (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene. 408:112–123.
- Cao SS, Du YZ. 2014. Characterization of the complete mitochondrial genome of Chilo auricilius and comparison with three other rice stem borers. Gene. 548:270–276.
- Chai HN, Du YZ, Zhai BP. 2012. Characterization of the complete mitochondrial genomes of Cnaphalocrocis medinalis and Chilo suppressalis (Lepidoptera: Pyralidae). Int J Biol Sci. 8:561–579.
- He SL, Zou Y, Zhang LF, Ma WQ, Zhang XY, Yue BS. 2015. The complete mitochondrial genome of the beet webworm, Spoladea recurvalis (Lepidoptera: Crambidae) and its phylogenetic implications. PLoS One. 10:e0129355.
- Ye F, Shi YR, Xing LX, Yu HL, You P. 2013. The complete mitochondrial genome of Paracymoriza prodigalis (Leech, 1889) (Lepidoptera), with a preliminary phylogenetic analysis of Pyraloidea. Aquat Insects. 35:71–88.