Abstract
In this study, DNA barcodes were generated for 40 species belonging to 32 genera under 10 families of Ephemeroptera from South India. Nucleotide sequence divergences were calculated using the Kimura two-parameter distance model and a neighbour-joining analysis was performed to provide a graphic display of the patterns of divergence among the species. This study demonstrates that COI barcoding is effective in discriminating among the mayfly species of South India, and provides a reference library for their future molecular identification.
Mayflies are an archaic lineage of insects, dating back to the late Carboniferous or early Permian periods, some 290 mya (Brittain & Sartori Citation2003). They occupy freshwater and brackish water habitats across the world, with the exception of Antarctica. They constitute an important part of the food chain, mainly consuming primary producers such as algae and plants, and as a food source for vertebrate predators like fish. They are excellent biological indicators of water quality and habitat quality (Sivaramakrishnan et al. Citation1996; Buffagni Citation1997; Selvakumar et al. Citation2014). They are ideal objects for integrated phylogenetic, biogeographic and phylogeographic studies, being endowed with several archaic traits in all life stages along with rather weak dispersal powers. Many of the montane mayflies, both nymphs and imagos are equally charismatic. Nymphs are important for freshwater ecological and biomonitoring studies, but difficulties in their species identification level impede research.
DNA barcoding can contribute to speeding up local biodiversity assessments to prioritise conservation areas or to evaluate the success of conservation actions and provide information about evolutionary histories (Krishnamurthy & Francis Citation2012). The application of DNA barcoding to freshwater biomonitoring has generated much interest for several reasons (Hajibabaei et al. Citation2011; Pilgrim et al. Citation2011; Sweeney et al. Citation2011). DNA barcodes have also implied in studying the systematics, diversity, ecology, biogeography, and conservation of aquatic insects (Sivaramakrishnan et al. Citation2014; Gattolliat et al. Citation2015). A comprehensive barcode library has been established for mayflies from Canada, Mexico, and the United States (Ball et al. Citation2005; Zhou et al. Citation2009, Citation2010; Webb et al. Citation2012; Gattolliat et al. Citation2015). To our knowledge, no molecular work of this kind was undertaken on mayflies in India so far. The emerging trends in molecular systematics and molecular phylogeny of mayflies are quite evident from the review by Sivaramakrishnan et al. (Citation2011). Our general aim is to develop a strategy for rapid construction of regional barcode libraries, and specific aim is to examine the efficiency of DNA barcoding for differentiating morphospecies. Present study deals with nymphs of mayflies due to their importance in freshwater ecology and for their biomonitoring value.
Mayfly nymphs were collected from stream and river basins of South India. The collected specimens were identified using scattered Indian mayfly taxonomic literature, under a stereo-zoom microscope. Samples used in this study included 44 specimens representing 40 species belonging to 32 genera and 10 families of Ephemeroptera from South India. Thirty-eight species were represented by single specimens, and 2 species characterized by more than one specimens (). DNA was extracted using DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). The mtCOI gene was amplified using universal primer LC01490 and HC02198 (Folmer et al. Citation1994). Sequencing was performed commercially by Amnion Biosciences Pvt. Ltd (Bangalore, India). Forward and reverse sequencing reads were assembled and corrected using BioEdit (Carlsbad, CA) and aligned using CLUSTALW (Cambridgeshire, UK). Neighbour-joining (NJ) tree and intraspecific and interspecific genetic divergence values were performed based on the Kimura 2-parameter (K2P) model using MEGA 5 (Tamura et al. Citation2011).
Table 1. Details of sample used in this study.
The present study established DNA barcode for 40 species of mayflies from South India through Genbank and BOLD systems. This is the first report of DNA barcode to the 40 species of mayflies from South India. Species details and sequence and barcode information are available at BOLD Systems (www.barcodinglife.org, ‘Molecular characterization of South Indian mayflies’ project). The details of the species along with their GenBank Accession numbers and Barcode ID are given in . Mean interspecific divergences computed for 40 species of South Indian mayflies ranged from 0.143% to 0.466%. The mean of all interspecific divergences was computed as 0.301%. The low levels of interspecific divergence occurred between two species within a genus and between the genus, such as Nathanella indica and Nathanella saraswathiae (0.17%), Cloedes soldani and Labiobaetis soldani (0.18%), Labiobaetis jacobusi and Cloeon bicolor (0.19%), and Choroterpes nambiyarensis and Indialis badia (0.14%).
Relatively, intraspecific genetic divergences were observed in the branches corresponding to species complexes in the NJ tree, as in Petersula courtallensis, which were divided into two subclades. The genetic divergence ranged from 0.003% to 0.051%, suggesting that more than one species will be represented. There was no difference between the closely related species Labiobaetis jacobusi and L. soldani. Three species, namely Baetis sp., Caenis sp., and Procloeon sp. were morphologically very distinct and also the present barcoding study clearly distinguished from their closely related species. The results of the overall NJ analysis of distances among the samples of 40 species are summarized in . The obtained results indicate that the portion of COI used as a DNA barcode effectively discriminates among mayfly species. It should be noted that the tree presented here is intended as a representation of the distance matrix only, and should not be interpreted as a phylogenetic hypothesis.
Figure 1. A Kimura 2-parameter NJ tree showing the DNA barcoding profile for 44 specimens of 40 nominal mayfly species from South India.
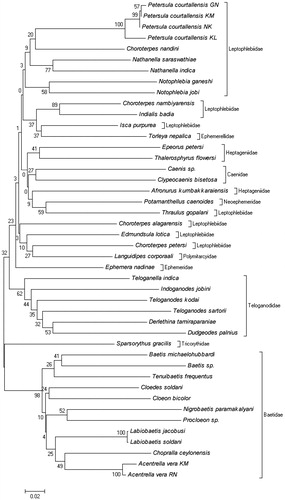
The present study reports for the first time COI barcode sequences for the purpose of species identification and the basis of global biodiversity assessment. All the species gave distinct COI sequences except Labiobaetis jacobusi and L. soldani, distinguishing them from conspecifics through the DNA barcode method (). Detailed molecular analysis is required to differentiate Labiobaetis jacobusi and L. soldani using more samples. Minimum level of intraspecific genetic divergence were found in Acentrella vera (0.022%), though it is distributed over a very wide area within the Oriental Realm (Kluge et al. Citation2014). Maximum level of intraspecific genetic divergence was found in Petersula courtallensis ranging from 0.003% to 0.051%. In order to confirm this and to describe a new species, it will be necessary to perform detailed morphological and molecular studies. The possibility of the presence of cryptic species complex within the genus Petersula may not be ruled out. However, further detailed investigations are necessary to understand clearly the taxonomic situation of Labiobaetis species and Petersula courtallensis. Baetis sp., Caenis sp., and Procloeon sp. were very distinct species based on this barcoding study. It will be required to perform thorough morphological studies to describe the valid species. The NJ tree supported the results of previous studies that have found the COI barcode to be an effective tool for the identification in mayflies (Ball et al. Citation2005; Zhou et al. Citation2010).
The present study confirms that, DNA barcode can be used effectively for species identification of South Indian mayfly species although the success rates vary with the level of genetic structure and demographic history. DNA barcoding analysis represents an interesting approach to new studies of taxonomy and species recognition of South Indian mayflies as new species, including cryptic species. Also, DNA barcoding can be used to analyze a mayfly community to estimate species richness of an entire mayfly community and for further phylogeographic studies. The results indicate that more taxonomic and molecular work are required on Indian Ephemeroptera as many currently recognized species include several highly divergent, often polyphyletic, haplotypes, usually correlated with morphological differentiation among lineages.
Acknowledgements
The authors gratefully acknowledge the Department of Zoology, University of Madras, for providing assistance through the UGC-SAP.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding
C. Selvakumar thanks the University Grants Commission (UGC), New Delhi, India, for award of Dr. D. S. Kothari Post Doctoral Fellowship [No.F.4-2/2006 (BSR)/13-670/2012 (BSR)].
References
- Ball SL, Hebert PDN, Burian SK, Webb JM. 2005. Biological identifications of mayflies (Ephemeroptera) using DNA barcodes. J North Am Benthol Soc. 24:508–524.
- Brittain JE, Sartori M. 2003. Ephemeroptera (mayflies). In: Resh VH, Carde RT, editors. Encyclopedia of insects. New York: Academy Press; p. 373–380.
- Buffagni A. 1997. Mayfly community composition and the biological quality of streams. In: Landolt P, Sartoti M. editors. Ephemeroptera and Plecoptera: Biology-Ecology-Systematics. MTL Fribourg: Bulletin de I’Institut Royale des Sciences Naturelles de Belgique; p. 235–236.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech. 3:294–299.
- Gattolliat JL, Cavallo E, Vuataz L, Sartori M. 2015. DNA barcoding of Corsican mayflies (Ephemeroptera) with implications on biogeography, systematics and biodiversity. Arth Syst Phyl. 73:3–18.
- Hajibabaei M, Shokralla S, Zhou X, Singer GAC, Baird DJ. 2011. Environmental barcoding: a next-generation sequencing approach for biomonitoring applications using river benthos. PLoS One. 6:e17497.
- Kluge NJ, Sivaramakrishnan KG, Selvakumar C, Kubendran T. 2014. Notes about Acentrella (Liebebiella) vera (Müller-Liebenau, 1982) (= Pseudocloeon difficilum Müller-Liebenau, 1982 syn. n. = Platybaetis arunachalae Selvakumar, Sundar, and Sivaramakrishnan, 2012 syn. n.) (Ephemeroptera: Baetidae). Aquatic Insects. 35:63–70.
- Krishnamurthy PK, Francis RA. 2012. A critical review on the utility of DNA barcoding in biodiversity conservation. Bio Conserv. 21:1901–1919.
- Pilgrim EM, Jackson S, Swenson S, Turcsanyi I, Friedman E, Weigt L, Bagley MJ. 2011. Incorporation of DNA barcoding into a large-scale biomonitoring program: opportunities and pitfalls. J North Am Benthol Soc. 30:217–231.
- Selvakumar C, Sivaramakrishnan KG, Janarthanan S, Arumugam M, Arunachalam M. 2014. Impact of riparian land use patterns on Ephemeroptera community structure in river basins of southern Western Ghats, India. Knowl Manag Aqua Ecosys. 412:1–15.
- Sivaramakrishnan KG, Janarthanan S, Selvakumar C, Arumugam M. 2014. Aquatic insect conservation – a molecular genetic approach. Conserv Genet Res. 6:849–855.
- Sivaramakrishnan KG, Morgan HJ, Vincent RH. 1996. Biological assessment of the Kaveri river catchment, South India, and using benthic macroinvertebrates: applicability of water quality monitoring approaches developed in other countries. Int J Ecol Environ Sci. 32:113–132.
- Sivaramakrishnan KG, Subramanian KA, Arunachalam M, Selva-kumar C, Sundar S. 2011. Emerging trends in molecular systematics and molecular phylogeny of mayflies (Insecta: Ephemeroptera). J Threat Taxa. 3:1972–1980.
- Sweeney BW, Battle JM, Jackson JK, Dapkey T. 2011. Can DNA barcodes of stream macroinvertebrates improve descriptions of community structure and water quality? J North Am Benthol Soc. 30:195–216.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Webb JM, Jacobus LM, Funk DH, Zhou X, Kondratieff B, Geraci, CJ, DeWalt RE, Baird DJ, Richard B, Phillips I, Hebert PDN. 2012. A DNA Barcode library for North American Ephemeroptera: progress and prospects. PLoS One. 7:e38063.
- Zhou X, Adamowicz SJ, Jacobus LM, DeWalt RE, Hebert PDN. 2009. Towards a comprehensive barcode library for arctic life – Ephemeroptera, Plecoptera, and Trichoptera of Churchill, Manitoba, Canada. Front Zool. 6:30.
- Zhou X, Jacobus LM, DeWalt RE, Adamowicz SJ, Hebert PDN. 2010. Ephemeroptera, Plecoptera, and Trichoptera fauna of Churchill (Manitoba, Canada): insights into biodiversity patterns from DNA barcoding. J North Am Benthol Soc. 29:814–837.
