Abstract
Sclerotinia sclerotiorum is one of the most devastating necrotrophic fungal plant pathogens in agriculture causing diseases in over 400 species of plants including important crops and numerous weeds. In this work, the mitochondrial sequence of S. sclerotiorum with different strain obtained from the infected stems of Brassica campestris L. in Wangjiang County, Anhui Province, China is presented. The mt DNA codes for 14 proteins of the respiratory chain, 1 ribosomal protein, 2 homing endonucleases, 2 rRNAs, 25 tRNAs, and 5 hypothetical proteins ORFs. Phylogenetic analysis with protein-coding gene sequences of reported Ascomycota mt genomes revealed the close relationship of JX-21 with the family of Sclerotiniaceae.
Sclerotinia sclerotiorum JX-21 (Fungi, Ascomycota, Ascomycetae, Helotiales, Sclerotiniaceae), is one of the most devastating necrotrophic fungal plant pathogens in agriculture (Bary Citation1886). It can cause diseases in over 400 species of plants including important crops and numerous weeds (Boland & Hall Citation1994). The majority of these species are dicotyledonous, such as sunflower, soybean, edible dry bean, and so on, although a number of agriculturally significant monocotyledonous plants are also hosts such as onion and tulip (Bolton et al. Citation2006). In recent years, 46 Pezizomycotina strains have been sequenced successfully. Most of them are available for members of the Eurotiomycetes and Sordariomycetes with only partial non-annotated genomes or draft annotations available for helotialean species (Duò et al. Citation2012). Only three mitochondrial genomes belong to members of the Sclerotiniaceae: S. sclerotiorum, S. borealia (Mardanov et al. Citation2014), and Botryotinia fuckeliana. Here, we resequenced the complete nucleotide sequence of the mt genome of S. sclerotiorum with different strains isolate obtained from the infected stems of Brassica campestris L., which was collected from March to April in 2012 from Wangjiang County (30°13′ N and 116°68′ E), Anhui Province, China and the strain was stored in the Chinese common microbe bacterial preservation administration centre with the preservation number CGMCC3.18027. The samples were processed within 24 h following the collection and were identified using morphological analysis and ITS sequencing (Guo et al. Citation2000). The DNA library was sequenced on an IlluminaMiseq in Majorbio and assembled with SOAP denovo v2.04 (Shenzhen, China). The mitochondrial genome of S. sclerotiorum strain JX-21 was shown to be a circular DNA molecule of 83,937bp (GenBank KX351425) with an average G + C content of 30.6%. All coding genes are transcribed with different polarities and started with the canonical translation initiation codon-AUG. Fourteen of the 15 typical mitochondrial protein-coding genes are involved in energy and oxidative metabolism (cox1, cox2, and cox3, cob, atp6, atp8, atp9, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6). The remaining one encoded the 40S ribosomal protein S3 (rps3).
Fourteen intronic ORFs were located in the five protein-coding genes cox1 (7), nad5 (3), nad1 (1), atp6 (1), cox3 (1), and rnl (1). Ten of the 14 ORFs exhibited similarity in the amino acid sequence to the LAGLIDADG motif and four of them were similar to GIY-YIG motif.
Twenty-five tRNA genes were identified in S. sclerotiorum JX-21 mt DNA genome represented 19 amino acids except tRNACys and included three copies of tRNAMet, tRNASer, and two copies of tRNAAla, tRNAThr. Single-copy genes encoded the remaining tRNAs. Nineteen tRNAs were grouped into four clusters with 6, 5, 3, and 5 tRNA genes each. Two clusters of 6 and 5 tRNAs were located around cox1 gene. The remaining two clusters were located around rRNA gene: five around rnl and three around rps3, while the remaining four tRNA genes occurred, respectively.
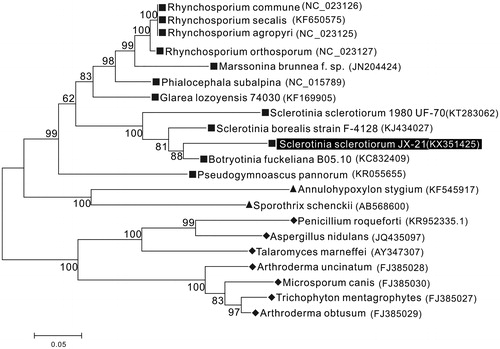
The phylogenetic analysis of S. sclerotiorum JX-21 was performed by comparison with 15 core mt proteins of other 21 species in Ascomycota (Mardanov et al. Citation2014) and was constructed by a maximum-likelihood analysis of MEGA 6.0 (Hachioji, Tokyo, Japan) (Tamura et al. Citation2013) program using 1000 bootstrap replicates. Result shows JX-21 is closely related to S. sclerotiorum1980 UF-70, S. borealia F-4128, and B. fuckeliana B05.10 which are another three members in the family Sclerotiniaceae with high bootstrap value supported (). In conclusion, the complete mtDNA of S. sclerotiorum JX-21 provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis for Sclerotiniaceae.
Figure 1. Molecular phylogenetic analysis of S. sclerotiorum JX-21 and other Ascomycotina species using the amino acid sequences of 15 common protein-coding genes and the maximum-likelihood method based on the Whelan and Goldman (WAG) model. Numbers at branch nodes are percentages based on 1000 bootstrap resampling. Three classes of filamentous ascomycetes are clearly distinguished as monophyletic groups: square-Leotiomycetes (represented by Helotiales), triangle-Sordariomycetes, and rhombus-Eurotiomycetes. Complete mitogenomes of other species are downloaded from NCBI and the GenBank accession numbers are given in the bracket after the species name.
Disclosure statement
We extend our thanks to the reviewers for their careful reading and helpful comments on this manuscript. The authors report no declarations of interest. The authors alone are responsible for the content and writing of the manuscript.
References
- Bary A. 1886. Ueber einige sclerotinien und sclerotienkrankheiten. Bot Zeit. 44:377 et seq. [M].
- Boland G, Hall R. 1994. Index of plant hosts of Sclerotinia sclerotiorum. Can J Plant Pathol. 16:93–108.
- Bolton MD, Thomma BP, Nelson BD. 2006. Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol Plant Pathol. 7:1–16.
- Duò A, Bruggmann R, Zoller S, Bernt M, Grünig CR. 2012. Mitochondrial genome evolution in species belonging to the Phialocephala fortinii sl-Acephala applanata species complex. BMC Genomics. 13:1.
- Guo L, Hyde K, Liew E. 2000. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol. 147:617–630.
- Mardanov AV, Beletsky AV, Kadnikov VV, Ignatov AN, Ravin NV. 2014. The 203 kbp mitochondrial genome of the phytopathogenic fungus Sclerotinia borealis reveals multiple invasions of introns and genomic duplications. PLoS One. 9:e107536.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
