Abstract
Lycodon liuchengchaoi is a new species discovered in recent years which is widely distribute in Anhui, Hubei and Sichuan Province. In this study, we determined the complete mitochondrial genome of L. liuchengchaoi. The result shows that the complete mitogenome of L. liuchengchaoi is 17,171bp. It is similar with the typical mtDNA of Serpentes, which contains 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, 2 control regions, and a stem-loop region. The phylogenetic tree, contains 17 Serpentiforms species, is divided into two clades which correspond to six genera in Colubridae. The L. liuchengchaoi which appeared into Clade A, clustered within Lycodon.
The Lycodon liuchengchaoi is a species of Colubridae family (Zhang et al. Citation2011). Lycodon liuchengchaoi, once misidentified as L. fasciatus because of their similarity in shape (Lei et al. Citation2014; Ding et al. Citation2015). However, in 2011, it was identified as a new species after the careful examination of these specimens that was collected from Sichuang Province (Zhang et al. Citation2011), the researchers noticed that they can be distinguished from L. fasciatus by several morphological characters, such as the anal plate and the large number and yellow colour of the rings around the body (Vogel et al., Citation2010; Zhang et al. Citation2011). In the following research, some new records about L. liuchengchaoi were reported in Anhui and Hubei Province (Zhang et al. Citation2015). In the present study, the L. liuchengchaoi sample was collected from Yaoluoping, Anqing of Anhui province, China (N 30°57′57.06″, E 116°04′04.96″). Now, the specimen was deposited in the laboratory of Evolution and Ecology, School of Life Sciences, Anhui University.
We sheared the muscle tissue to extract the whole genomic DNA using a standard proteinase-K/phenol–chloroform protocol (Sambrook & Russell Citation2006). The entire mitogenome was amplified using 16 pairs of primers by PCR. Here, we sequenced the complete mitochondrial genome of L. liuchengchaoi and submitted it to GenBank (accession no. KX553922). Phylogenetic relationship of 17 Colubridae species were analyzed with the Bayesian inference (BI) method using the MrBayes version 3.1.2 software (University of Rochester, NY) (Huelsenbeck & Ronquist Citation2001) based on nucleotide sequences of Cyt b gene, selecting Python molurus as an outgroup. In this process, the best-fitting nucleotide substitution model (GTR + I + G) was selected via MrModeltest version 2.1 (Uppsala University, SWE) (Nylander et al. Citation2004); the four independent Markov’s chain runs for 1,000,000 metropolis-coupled Monte Carlo (MCMC) generations, sampling every 1000 generations. When the average standard deviation of split frequencies reached a value less than 0.01, the first 1000 trees were discarded as ‘burn-in’ and the remaining trees were used to calculate the Bayesian posterior probabilities (Pan et al. Citation2015).
The complete mitogenome sequence of L. liuchengchaoi is 17,171 bp in length, and the gene order was identical to L. semicarinatum, including 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, 2 control regions, and a stem-loop region. The base composition of the mitogenome was 33.8% A, 28.2% C, 13.0% G, and 25.0% T. The ND6 subunit gene, stem-loop, and eight tRNA genes (tRNAPro, tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), and tRNAGlu) were encoded on the L-strand, the others were encoded on the H-strand.
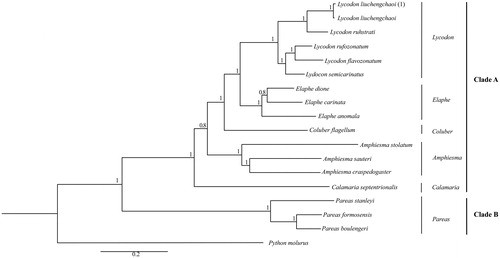
According to the Bayesian analysis, the phylogenetic tree divided the species into two clades, these relationships received well support (). The Clade A contains Lycodon, Elaphe, Coluber, Calamaria, and Amphiesma. The Pareas presents in the Clade B. In the phylogenetic tree, the species of sample in this study named as L. liuchengchaoi (1) cluster with the sequence of L. liuchengchaoi (Zhang et al. Citation2015), which appeared in the genus of Lycodon, Clade A. Our study of L. liuchengchaoi can provide a useful database for analyzing the classification and status in Colubridae.
Figure 1. The phylogenetic relationships analysis among the species in Colubridae is based on the Cyt b gene using Bayesian inference (BI). The percentages of Bayesian’s posterior probabilities (BPPs) show as the numbers at each node. GenBank accession numbers Cyt b are: L. liuchengchaoi (KP898899), L. ruhstrati (NC029153), L. rufozonatum (NC028730), L. flavozonatum (NC028730), L. semicarinatus (NC001945), E. dione (HQ830257), E. carinata (KF669252), E. anomala (KP900218), C. flagellum (KM403637), A. stolatum (KJ685711), A. crapedogaster (GQ281178), A. sauteri (AF402905), C. septentrionalis (KR814699), P. stanleyi (JN230704), P. formosensis (JF827693), P. boulengeri (JF827683) and P. molurus (AY099983).
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
References
- Ding M, Wu J, Qian L, Pan T, Zhang BW. 2015. Complete mitochondrial genome of Lycodon flavozonatum and implications for Colubridae taxonomy. Mitochondrial DNA. 27:3473–3474.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Lei J, Sun X, Jiang K, Vogel G, Booth DT, Ding L. 2014. Multilocus phylogeny of Lycodon and the taxonomic revision of Oligodon multizonatum. Asian Herpetol Res. 5:26–37.
- Nylander JA, Ronquist F, Huelsenbeck JP, Nievesaldrey J. 2004. Bayesian phylogenetic analysis of combined data. Syst Biol. 53:47–67.
- Pan T, Wang H, Hu C, Sun ZL, Zhu X, Meng T, Meng X, Zhang BW. 2015. Species delimitation in the genus moschus (Ruminantia: Moschidae) and its high-plateau origin. PLoS One. 10:e0134183.
- Sambrook J, Russell DW. 2006 . Purification of nucleic acids by extraction with phenol: chloroform. CSH Protoc. 2006:pdb. prot4455.
- Vogel G, David P, Pauwels O, Sumontha M, Norval G, Hwedrix R, Vu N, Ziegler T. 2010. A revision of Lycodon ruhstrati (Fischer 1886) auctorum (Squamata Colubridae), with the description of a new species from Thailand and a new subspecies from the Asian mainland. Tropical Zool. 22:131–182.
- Zhang J, Jiang K, Vogel G, Rao D. 2011. A new species of the genus Lycodon (Squamata, Colubridae) from Sichuan Province, China. Zootaxa. 2982:59–68.
- Zhang L, Li FP, Lei Y, Zheng PW, Huang LQ, Huang S. 2015. New Record of Lycodon liuchengchaoi in Anhui. Zool Res. 36:178–180.
