Abstract
Pacific golden plover Pluvialis fulva (Charadriiformes, Scolopacidae) is obligate long-distance migrant bird breeding from northernmost Asia into western Alaska, and wintering on islands in the Pacific Ocean and Australia. In this study, we report the complete mitochondrial genome of P. fulva, which is a circular molecule of 16,854 bp in size and consists of 13 protein-coding genes, 2 ribosomal RNAs, 22 transfer RNAs, and a control region. The A + T content of the overall base of the composition of H-strand is 54.88% (A: 31.48%, T: 23.40%, C: 31.48%, and G: 13.65%). It is very interesting that there are some insertions/deletions in the cytb gene sequence (total of 10bp deletions and 5bp insertions in four positions), which may result in reading frame shift and/or internal stops. Some short microsatellite-like repeat regions (ACCACCC) are scatter in the control region. The phylogenetic analysis resolved a well-supported clade of Charadriiformes in which Pluvialis appears to be sister group of Vanellus. This mitogenome provides a valuable resource for further study of molecular systematics, species identification, population genetics, phylogeography, and conservation genetics of Charadriiformes.
Keywords:
Pacific golden plover Pluvialis fulva (Charadriiformes, Scolopacidae) is strongly migratory, with different populations travelling on narrow or broad front. Despite the fact that the population trend appears to be decreasing, it is evaluated as Least Concern (BirdLife International Citation2012). Many studies on morphology, ecology, behaviour, bioenergetics, and migration had been reported (Johnson et al. Citation2001, Citation2009; Withrow & Winker Citation2014; Jukema et al. Citation2015), but the basic genetics data are relatively unclear, and the taxonomic assignment has been debated recently (Baker et al. Citation2012; Withrow & Winker Citation2014). In this study, we sequenced the complete mitochondrial genome of P. fulva.
The complete mitogenome of P. fulva has been sequenced to better understand the mitogenomic characteristics and its phylogenetic relationships within Charadriiformes. The muscle specimen of P. fulva was collected from the costal of Rudong Country, Jiangsu Province, China (32°32′43.42″ N, 121°06′09.02″ E). A voucher specimen was preserved in absolute ethanol at Nanjing Normal University (NJNU: Pfu-2015007), Nanjing, China. Total DNA was extracted with standard phenol–chloroform methods according to Sambrook and Russell (Citation1989).The complete mitochondrial genome was amplified and sequenced by 13 pairs of primers. Annotations were confirmed by comparing 17 Charadriiformes species with MITOS-generated annotations (Bernt et al. Citation2013).
The circular genome is 16,854 bp in length with 13 protein-coding genes, 2 ribosomal RNAs (12S rRNA and 16S rRNA), 22 transfer RNA genes, and a noncoding region. The overall nucleotide composition was A: 31.48%, T: 23.40%, C: 31.48%, and G: 13.65%. The annotated mitogenome of P. fulva is available online in NCBI (GenBank accession number: KX639757). It is very interesting that there are some insertions/deletions in the cytb gene sequence (total of 10bp deletions and 5bp insertions in four positions) which may result in reading frame shift and/or internal stops.
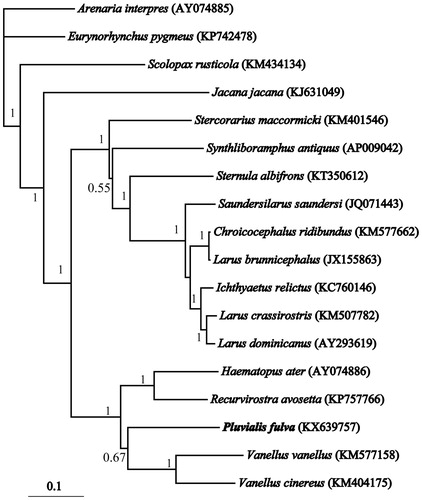
Seventeen closely related Charadriiformes mitogenomes (82.37%–86.43% identical bases) were downloaded and aligned with Clustal X 1.81 (Thompson et al. Citation1997; Strasburg, Alsace, France) using default parameters. There were 6572 (57.59%) conserved sites, 3612 (31.65%) parsim-info sites, and 1213 (10.63%) singleton sites. Total of 11,412 bp (with the concatenated 13 protein-coding genes) were used for phylogenetic analyses. The Bayesian phylogenetic analyses were performed using MrBayes 3.1.2 (Ronquist et al. Citation2012). Models of molecular evolution were assessed using MrModeltest 2.3 (Nylander Citation2004). The Four Markov Chains Monte Carlo (MCMC) chains were run for 1.0 × 106 generations. Two independent runs were performed to allow additional confirmation of the convergence of MCMC runs.
The phylogenetic analysis () resolved a well-supported clade of Charadriiformes in which Pluvialis appears to be a sister group of Vanellus. The result indicated that there was great mitochondrial divergence within the Charadriiformes. The newly described mitogenome can be used in the future to disentangle the taxonomy within Plover, and provided a valuable resource for the studies related to shorebird’s molecular population genetics, phylogeography, and systematics.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
References
- Baker AJ, Yatsenko Y, Tavares ES. 2012. Eight independent nuclear genes support monophyly of the plovers: the role of mutational variance in gene trees. Mol Phylogenet Evol. 65:631–641.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendor M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- BirdLife International. 2012. Pluvialis fulva. The IUCN Red List of Threatened Species 2012: e.T22693735A38568056. Downloaded on 6 August 2016.
- Johnson OW, Adler CD, Ayres LA, Bishop MA, Johnson PM. 2009. Radio-tagged pacific golden-plovers: further insight concerning the hawaii-alaska migratory link. Wilson Bull. 116:158–162.
- Johnson OW, Bennett AJ, Iii LA, Bennett LA, Johnson PM, Morgart JR, Kienholzg RJ. 2001. Radio-tagged pacific golden-plovers: the Hawaii-alaska link, spring destinations, and breeding season survival. J Field Ornithol. 72:537–546.
- Jukema J, Rhijn JGV, Piersma T. 2015. Geographic variation in morphometrics, molt, and migration suggests ongoing subspeciation in pacific golden-plovers (Pluvialis fulva). AUK. 132:647–656.
- Nylander JAA. 2004. MrModeltest v2. Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University.
- Ronquist F, Teslenko M, van der Mark P, Ayres DJ, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biol. 61:539–542.
- Sambrook J, Russell DW. 1989. Molecular cloning: a laboratory manual. vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882.
- Withrow JJ, Winker K. 2014. Genetics of a high-latitude cryptic speciation event: American and Pacific golden-plovers. Wilson J Ornithol. 126:429–442.