Abstract
Chrysomya phaonis (Seguy, 1928) is one of the blowflies of great medical and forensic importance. In this paper, we report that the entire genome of mitochondrial DNA of C. phaonis is 15,831 bp in length, which consists of 39 genes including13 protein-coding genes, 24 tRNA genes, 2 mitochondrial ribosomal RNA genes, and a 992 bp non-coding A + T-rich region. The overall base compositions of A, G, C, and T are 38.79%, 9.75%, 14.15%, and 37.31%, respectively. We provide the first complete mitochondrial genome of C. phaonis, and should provide useful information for phylogenetic and species identification for C. phaonis.
Blowflies (Diptera: Calliphoridae) are distributed worldwide, some species play important role in the mechanical transfer of disease to human beings and animals (Norris Citation1965; Zumpt Citation1965; Greenberg Citation1971; Greenberg Citation1973; Kuhlhorn Citation1983; Ghandour Citation1988). Chrysomya phaonis (Seguy, 1928) is one of the most important oriental species of the blowflies, C. phaonis distributes in China, India, Nepal, and Afghanistan (Fan Citation1992; Fan et al. Citation1997; Kurahashi et al. Citation1994). However, there is limited molecular biology information about this species. Viewing it is a potential medical vector and indictor in forensic science, we report here the complete mitochondrial genome of C. phaonis for species identification and phylogenetic analysis.
Exampled sample of C. phaonis were obtained in Bayi County, Tibet, China (N29°37′31.39″; E94°23′25.01″) in July 2013. The studied specimen is stored in the medical vector collections of Zhongshan Entry-Exit Inspection and Quarantine technology center, and the accession number to the specimen is 20130728-232F-Baiyi. We designed 10 pairs of oligo-nucleotide primers according to the conserved regions from reported mitochondria genome sequences of its most related species C. pinguis.
The complete mitochondrial genome of C. phaonis (GenBank accession KX500359) is 15,831 bp in length, which consists of 39 genes () including 13 protein-coding genes, 24 tRNA genes, 2 mitochondrial ribosomal RNA genes (12S rRNA and 16S rRNA), and a 992 bp non-coding A + T-rich region. The 13 protein-coding genes include seven NADH dehydrogenase (ND1-6 and ND4L), three subunits of cytochrome oxidase (COI-III), two subunits of ATP synthase (ATP4 and ATP6), one subunit of cytochrome b (Cytb). Twelve of the 13 protein-coding genes were identified with ATN as start codon coding for M except for COI, which is similar to the former studies (Yan et al. Citation2014; Zhong et al. Citation2016).
Table 1. Mitochondrial gene profile of C. phaonis.
Chrysomya phaonis not only could cause harm to human health as medical vectors (Norris Citation1965; Zumpt Citation1965; Greenberg Citation1971; Kuhlhorn Citation1983; Ghandour Citation1988), but also are significant in forensic science (Harvey et al. Citation2008). DNA typing of forensic insect specimens offers a quick and reliable alternative, so more and more researchers have started using the whole sequences of mitochondrial genome or part of the genes to identify species of Blowflies (Wells & Williams Citation2007; Harvey et al. Citation2008; Desmyter & Gosselin Citation2009; DeBry et al. Citation2013).
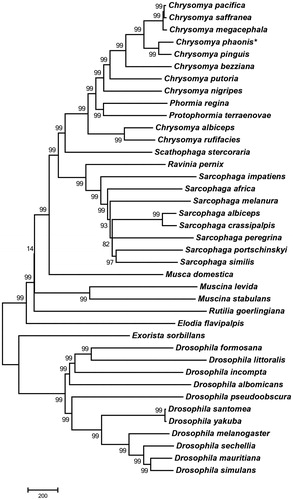
Here, we provide the entire genome of mitochondrial DNA of C. phaonis. The phylogenetic analysis of C. phaonis was performed by comparison with other 39 Diptera species mitochondrial genomes (). Phylogenetic tree was generated by a neighbour-joining analysis of MEGA 6.0 program (Tamura et al. Citation2013) using 1000 bootstrap replicates. The N-J tree revealed that C. phaonis was placed mostly close to C. pinguis, which could not be distinguished with the common adopted COI DNA barcode sequences. So, it may provide some help for the molecular identification of C. phaonis, in particular to distinguish C. phaonis from its closely related species C. pinguis.
Figure 1. The neighbour-joining (NJ) tree of C. phaonis with other 39 Diptera species based on mitochondrial genomes. For each node, the bootstrap support was calculated using 1000 replicates. GenBank accession numbers of mitochondrial genomes used in this hylogeny analysis were listed: C. albiceps (NC_019631.1); C. bezziana (JX913737.1); C. megacephala (KT272775.1); C. nigripes (KT444441.1); C. pacifica (KP861632.1); C. pinguis (KM244730.1); C. putoria (AF352790.1); C. saffranea (JX913742.1); C. saffranea (JX913742.1); Protophormia terraenovae (JX913743.1); Sarcophaga africa (KM881633.1); S. albiceps (NC_028413.1); S. crassipalpis (KP861920.1); S. impatiens (NC_017605.1); S. melanura (NC_026112.1); S. peregrina (NC_023532.1); S. portschinskyi (NC_025574.1); S. similis (NC_025573.1); Scathophaga stercoraria (KM200724.1); Musca domestica (KT444442.1); Muscina stabulans (NC_029487.1); M. stabulans (NC_026292.1); Elodia flavipalpis (NC_018118.1); Exorista sorbillans (HQ322500.1); Ravinia pernix (NC_026196.1); Rutilia goerlingiana (NC_019640.1); Phormia regina (KC005712.1); Drosophila albomicans (NC_027937.1); Drosophila formosana (NC_028518.1); D. incompta (NC_025936.1); D. littoralis (FJ447340.1); D. mauritiana (AF200831.1); D. mauritiana (NC_005779.1); D. melanogaster (KT174474.1); D. pseudoobscura (NC_018348.1); D. santomea (KF824869.1); D. sechellia (AF200832.1); D. simulans (AY518674.1); D. yakuba (KF824899.1).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- DeBry RW, Timm A, Wong ES, Stamper T, Cookman C, Dahlem GA. 2013. DNA-based identification of forensically important Lucilia (Diptera: Calliphoridae) in the Continental United States. J Forensic Sci. 58:73–78.
- Desmyter S, Gosselin M. 2009. COI sequence variability between Chrysomyinae of forensic interest. Forensic Sci Int Genet. 3:89–95.
- Fan ZD. 1992. Key to the common flies of China. 2nd ed. Beijing (China): Science Press. p. 992.
- Fan ZD, Chen ZZ, Fang JM, Zheng SS, Tao ZL. 1997. Fauna Sinica Insecta: Vol. 6, Diptera: Calliphoridae. Beijing (China): Science Press. p. 186.
- Ghandour AM. 1988. Health hazards in humans and animals caused by imported livestock diseases in Saudi Arabia. Fauna Saudi Arabia. 9:468–477.
- Greenberg B. (1971). Flies and disease. In: Ecology, classification and biotic associations, vol. 1. Princeton, (NJ): Princeton University Press. p. 856
- Greenberg B. 1973. 447. Flies and diseases, biology and disease transmission. Princeton, (NJ): Princeton University Press.
- Harvey ML, Gaudieri S, Villet MH, Dadour IR. 2008. A global study of forensically significant calliphorids: implications for identification. Forensic Sci Int. 177:66–76.
- Kuhlhorn F. 1983. Verbreitung der Toxoplasmose. Katzenkot Und Dipteren. Tierarztl. Prax. 11:385–392.
- Kurahashi H, Thapa VK, Shinonaga S, Iwasa M. 1994. Notes on the Nepalese calliphorid flies (Insecta: Diptera). Jap J Sanit Zool. 45:179–252.
- Norris KR. 1965. The bionomics of blow flies. Annu Rev Entomol. 10:47–68.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Yan J, Liao H, Xie K, Cai J. 2014. The complete mitochondria genome of Chrysomya pinguis (Diptera: Calliphoridae). Mitochondrial DNA. http://dx.doi.org/10.3109/19401736.2014.958675.
- Wells JD, Williams DW. 2007. Validation of a DNA-based method for identifying Chrysomyinae (Diptera: Calliphoridae) used in a death investigation. Int J Legal Med. 121:1–8.
- Zhong M, Wang X, Liu Q, Luo B, Wu C, Wen J. 2016. The complete mitochondrial genome of the flesh fly, Boettcherisca peregrine (Diptera: Sarcophagidae). Mitochondrial DNA Part A. 27:106–108.
- Zumpt F. 1965. Myiasis in man and animals in the old world: a textbook for physicians veterinarians and zoologists. London (UK): Butterworth. p. 267.
