Abstract
The complete mitochondrial DNA genome of Terek sandpiper, Xenus cinereus, was detected by using polymerase chain reaction method and DNASTAR software package. The circular mitogenome (16,817 bp in length) contains 13 protein-coding genes, 2 rRNA genes (12S ribosomal RNA and 16S ribosomal RNA), 22 tRNA genes and a control region. The content of four kinds of bases of the complete mitochondrial DNA is 30.7% for A, 24.9% for T, 30.2% for C and 14.3% for G, respectively. To validate our data, 12 published complete mitochondrial genomes of Charadriiformes along with the genome of Terek sandpiper were used to construct the phylogenetic tree.
The Terek sandpiper, Xenus cinereus, is slightly larger than the common sandpiper and famous for its long upcurved bill, which makes it very distinctive. As the only member of the genus Xenus, the complete mitochondrial DNA of Terek sandpiper is essential to be explored so that we can gain insight into the phylogenetic relationship with other sandpiper. In recent years, the trend of population is stable, and the intriguing species is listed as Least Concern, but Terek sandpiper is still vulnerable somewhere due to the rapid development of habit, the loss of alternative intertidal wetland, the spread of invasive cordgrass Spartina alterniflora, pollution and increasing human disturbance (IUCN 2016) (Roderick & Stuart Citation2010) (Hua et al. Citation2015).
We collected the feather sample and a small amount of blood sample of Terek sandpiper when we banded large quantities of shorebirds on the eastern coast of Jiangsu province, China. The sample was preserved with ethanol in sterilized centrifuge tube and stored at −20 °C in the Nanjing Normal University. DNA was extracted by using traditional phenol–chloroform methods (Sambrook & Russell Citation1989).
After obtaining the original sequence data, the chromatograms of each sequence were proofread and assembled using Seqman Pro (DNASTAR, Inc., Madison, WI). The complete mtDNA genome has been uploaded to the GenBank (Accession Number: KX644890). The circular mitogenome is 16,817 bp in length, including 13 protein-coding genes, 2 rRNA genes (12S ribosomal RNA and 16S ribosomal RNA), 22 tRNA genes and a control region. A few intergenic spacers (ranging 1–15) and overlapping nucleotides (ranging 1–10) exist between different genes.
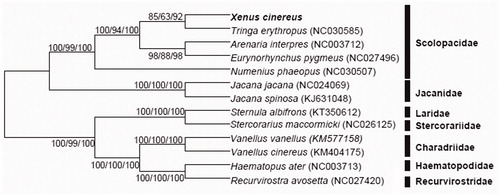
The Charadriiformes are distributed around the world, and contain 21 families with high-biodiversity. The complete mitochondrial genome of different species often unravel varied feature and can explain the evolution history. Here, 12 extra complete mitochondrial genomes were downloaded for constructing the phylogenetic tree with Terek sandpiper. All the sequences were aligned by Clustal X 2.1 software with default parameters and then modified manually (Thompson et al. Citation1997). Then, a total of 15,547 bp were used to construct the phylogenetic trees based on the maximum likelihood (ML), maximum parsimony (MP) and neighbour-joining (NJ) algorithm with MEGA 6 (Tamura K et al. Citation2013). The reliability of each phylogenetic tree was estimated by bootstrapping with 1000 bootstrap pseudosamples. The three phylogenetic trees were coincident with each other and the posterior probabilities shown above the branch indicate the relatively high reliability. The consensus tree demonstrates the Terek sandpiper is closely related to the Spotted redshank, which is classified into the family Scolopacidae (). We hope this study can help us comprehend the phylogenetic relationship within the Charadriiformes and illustrate the adaptive evolution of the Charadriiformes in the future.
Acknowledgements
We sincerely thank Dr. Chaochao Hu for his help and instruction. Meanwhile, we also thank our tutor Pro Chang, who encouraged us and provided us every facility.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding
This project was supported by grant from the National Natural Science Foundation of China (NSFC: 31071901) and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
References
- Hua N, Tan K, Chen Y, Ma Z. 2015. Key research issues concerning the conservation of migratory shorebirds in the yellow sea region. Bird Conserv Int. 25:38–52.
- IUCN. 2016. IUCN red list of threatened species (ver. 2016.1). Available from: http://www.iucnredlist.org [last accessed 31 July 2016].
- Roderick M, Stuart A. 2010. The status of threatened bird species in the Hunter Region. The Whistler. 4:1–28.
- Sambrook J, Russell DW. 1989. Molecular cloning: a laboratory manual, vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882.