Abstract
Shizothorax grahami (S. grahami) is an underlying economic cold-freshwater fish in southwest China. In this study, the complete mitochondrial genome of S. grahami was determined (GenBank accession number is NC_029708). The mitochondrial genome sequence of S. grahami was a circular molecule with 16,584 bp in length, and contained 37 typical animal mitochondrial genes including 2 ribosomal RNA genes, 13 protein-coding genes, 22 transfer RNA genes and a control region (D-loop). Four nucleotide compositions and their relative proportions of the entire mitogenome was 27.14% C, 19.93% G, 29.58% A and 25.34% T. Among most species of genus Shizothorax, Schizothorax davidi had the closest relationship with S. grahami, and then Schizothorax prenanti.
The Shizothorax grahami (S. grahami), an underlying economic cold-freshwater fish in southwest China, belongs to the family Cyprinidae and the order Cypriniformes (Si-Yu et al. Citation2012). It was mainly found in the lower stream of Jinshajiang River and its tributary the upper reaches of Wujiang River. In this study, we first determined the complete mitochondrial genome of S. grahami, which provides the basic molecular data for the further study on its systematics and conservation biology (Peng et al. Citation2006).
The complete mitochondrial genome of S. grahami was found to be a circular molecule with 16,584 bp in length, and has been deposited in GenBank database with Accession number of KU234535. The test fish sample was collected from Chishui River, Chishui City, Guizhou Province, China (28°28′45″N, 105°56′23″E). The specimens were immersed with formalde-hyde, and collected in the museum of Pearl River Fisheries Research Institute with accession number SC20151015004.
The DNA of S. grahami was extracted from the tail muscles using the salting-out procedure (Howe et al. Citation1997). We completely sequenced the genomic DNA of S. grahami using high-throughput sequencing technologies on the Illumina HiSeq2500 platforms (San Diego, CA), and the transfer RNA (tRNA) genes were identified using the tRNAscanSE 1.21 program (Schattner et al. Citation2005). As with other vertebrates (Boore Citation1999), mitochondrial genome of S. grahami contained 2 ribosomal RNA (rRNA) genes (12S rRNA and 16S rRNA), 13 protein-coding genes, 22 tRNA genes and a control region (D-loop). The overall base compositions of S. graham were C = 4501 (27.69%), G = 2974 (17.93%), A = 4906 (29.58%) and T = 4203 (25.34%). Except for NADH dehydrogenase subunit 6 (ND6) and eight tRNA genes which were encoded by the light (L)-strand, all the other mitochondrial genes of S. graham were encoded on the heavy (H)-strand (Pereira, Citation2000). Among the 13 protein-coding genes, all began with the ATG start codon except for cytochrome c oxidase subunit I (COX1) that started with GTG. Moreover, six protein-coding genes, including ND1, COX1, ND4L, ND5, ND6 and ATP synthase subunit 8 (ATP8), harbored the typical termination codon TAA or TAG, whereas the other seven genes, ND2, COX2, ATP6, COX3, ND3, ND4 and cytochrome b (CYTB), had an incomplete termination codon T or TA. As in the other vertebrates (Inoue et al. Citation2000), 12S rRNA and 16S rRNA were located between the tRNAPhe and tRNALeu genes and separated by the tRNAVal gene, at the length of 956 bp and2774 bp, respectively. The AT contents of the two rRNA genes were 51.15% and 56.41%, respectively. Twenty two tRNA genes were interspersed between rRNA genes and protein-coding genes, ranging in the length from 67 bp (tRNACys) to 76 bp (tRNALeu). The D-loop region (935 bp) was located between tRNAPro and tRNAPhe, with a high AT content of 66.20%.
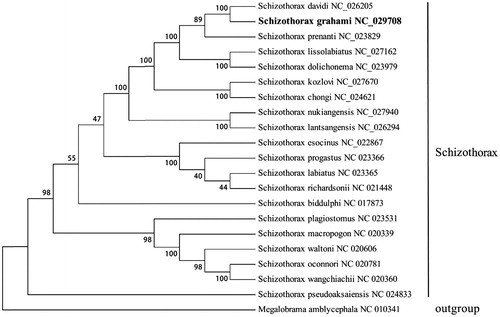
The phylogenetic tree was constructed on the basis of the complete mitochondrial genome sequences from S. grahami and the mitochondrial genome of the other 19 species of genus Shizothorax by the maximum parsimony method (). Numbers on the nodes correspond to bootstrap values based on 1000 iterations. The Megalobrama amblycephala was selected as outgroup. Among those 19 species of genus Shizothorax, Schizothorax davidi had the closest relationship with S. grahami, and then Schizothorax prenanti.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article. The authors have nothing to disclose.
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Howe J, Klimstra D, Cordon-Cardo C. 1997. DNA extraction from paraff in-embedded tissues using a salting-out procedure, a reliable method for PCR amplification of archiva1 material. Histol Histopathol. 12:595–601.
- Inoue JG, Miya M, Tsukamoto K, Nishida M. 2000. Complete mitochondrial DNA sequence of the Japanese sardine Sardinops melanostictus†. Fish Sci. 66:924–932.
- Peng Z, Wang J, He S. 2006. The complete mitochondrial genome of the helmet catfish Cranoglanis bouderius (Siluriformes: Cranoglanididae) and the phylogeny of otophysan fishes. Gene. 376:290–297.
- Pereira SL. 2000. Mitochondrial genome organization and vertebrate phylogenetics. Genet Mol Biol. 23:745–752.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Si-Yu HU, Zhan HX, Zhao HT, Chen YX. 2012. Artificial culture and breeding technique of Shizothorax grahami. Hubei Agricultural Sciences. 51:36–138. (in Chinese).