Abstract
The complete mitochondrial genome of Stegobium paniceum (Coleoptera: Anobiidae) is a circular DNA molecule of 15,271 bp (GenBank accession number XK819317), and its nucleotide composition is biased towards A + T nucleotides (78.32%). This genome comprises 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA (tRNA) genes, and an A + T-rich region. The gene order of S. paniceum was similar to those found in other known Coleoptera species. Sixteen reading frame overlaps and six intergenic regions were found in the mitochondrial genome of S. paniceum. All 22 tRNA genes have the typical cloverleaf secondary structure, with an exception for trnS1 (AGN). The phylogenetic relationships based on neighbour-joining method revealed that S. paniceum is closely related to Apatides fortis, which is consistent with the traditional morphological classification.
The drugstore beetle, Stegobium paniceum (Linnaeus) (Coleoptera: Anobiidae), is a major pest that cause tremendous damage to stored grains and seeds, packaged food products, animal-, and plant-derived products, including more than 300 drug materials (Li et al. Citation2009; Abdelghany et al. Citation2010). In this study, the S. paniceum samples were collected from medicinal materials company in Guizhou province of China (N26°31′, E106°43′). The specimen was deposited in Insect Collection (Accession Number GYU-Col-20090001-2), College of Biology and Environmental Engineering, Guiyang University, Guiyang, China.
The complete mitochondrial genome of S. paniceum (GenBank accession number XK819317) has been sequenced and annotated. It is a typical circular DNA molecule of 15,271 bp in length, larger than Eucryptorhynchus brandti (15,122 bp) and smaller than Tribolium castaneum (15,883bp), Dastarcus helophoroides (15,878 bp), and Hycleus chodschenticus (16,257 bp) (Liu et al. Citation2014; Zhang et al., Citation2014; Nan et al. Citation2016; Yuan et al. Citation2016).The nucleotide composition of mitochondrial genome of S. paniceum is heavily biased towards A + T nucleotides, accounting for A (41.62%), T (36.70%), G (9.06%), and C (12.62%). The AT-skew and GC-skew of this genome were 0.063 and −0.164, respectively.
The mitochondrial genome of S. paniceum contains 37 genes, including13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes. The gene order of S. paniceum was similar to those found in other known Coleoptera species. Gene overlaps 39 bp that have been found at 16 gene junctions, the longest 8 bp overlapping exists between trnW and trnC. The S. paniceum mitochondrial genome harbours six intergenic regions (trnS2 and nad1 share 20 nucleotides, trnM and nad2 share three nucleotides, nad4L and trnT share two nucleotide, trnY and cox1 share a nucleotide, trnP and nad6 share a nucleotide, and nad1 and trnL1 share a nucleotide). With an exception for trnS1 (AGN), all tRNAs have the typical cloverleaf secondary structure, which are common in most animal mitochondrial genome (Wolstenholme Citation1992). The length of these tRNAs vary from 62 bp for trnI, trnL1, trnY to 70 bp for trnM, A + T content ranged from 70.31% (trnF) to 90.48% (trnE).
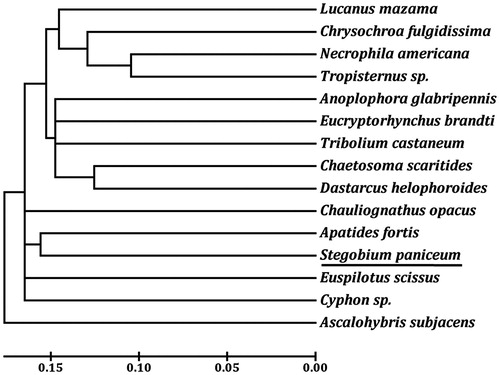
Two rRNA genes (rrnL and rrnS) are located between trnL1 and trnV, and between trnV and the A + T-rich region, respectively. The size of rrnL and rrnS of S. paniceum is 1254 bp and 755 bp long, respectively. Of the 13 PCGs, 11 genes start with ATN codons, including five ATAs (cox2, nad3, nad4, nad6, and nad2), four ATGs (atp6, cox3, nad4L, and cob), two ATTs (atp8 and nad5). However, cox1 and nad1 used AAA and TTG as start codon, respectively. Eight PCGs terminate with the conventional stop codons TAA (atp6, atp8, nad4L, nad6, and nad2) or TAG (nad3, cob, and nad1), the remaining PCGs including cox1, cox2, cox3, nad4, and nad5 use a single T as stop codon. The major A + T-rich region was located between rrnS and trnI genes with a length of 795 bp long, and the A + T content was 90.69%. Based on the concatenated amino acid sequences of 13 PCGs, the neighbour-joining method was used to construct the phylogenetic relationship of S. paniceum with 13 other representative beetles. The mitochondrial genome of neuropteran Ascalohybris subjacens (NC_011277) was used as an outgroup. The results demonstrated that S. paniceum is more closely related to Apatides fortis than other beetles (), which is consistent with the traditional morphological classification.
Figure 1. The neighbour-joining phylogenetic tree of S. paniceum and other beetles was constructed based on 13 mitochondrial protein-coding genes. The GenBank accession numbers used for tree constructed are as follows: A. fortis (NC_013582), Anoplophora glabripennis (NC_008221), Chaetosoma scaritides (NC_011324), Chrysochroa fulgidissima (NC_012765), Chauliognathus opacus (NC_013576), Cyphon sp. (NC_011320), D. helophoroides (NC_024271), E. brandti (NC_025945), Euspilotus scissus (NC_018353), Lucanus mazama (NC_80 013578), Necrophila americana (NC_018352), T. castaneum (NC_003081), and Tropisternus sp. (NC_018349).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding
This work was supported in part by the National Natural Science Foundation (31460476), the Science and Technology Foundation of Guizhou Province (QKHLH[2014]7167, QKHJZ[2014]2002), the National Training Program of Innovation and Entrepreneurship for Undergraduates (201510976038), International S&T Cooperation Program of Guizhou Province (QKHWG[2013]7047), and the Doctor Foundation Project of Guiyang University (GYU2014003).
References
- Abdelghany AY, Awadalla SS, Abdel-Baky NF, El-Syrafi HA, Paul GF. 2010. Effect of high and low temperatures on the drugstore beetle (Coleoptera: Anobiidae). J Econ Entomol. 103:1909–1914.
- Li C, Li ZZ, Cao Y, Zhou B, Zheng XW. 2009. Partial characterization of stress-induced carboxylesterase from adults of Stegobium paniceum and Lasioderma serricorne (Coleoptera: Anobiidae) subjected to CO2-enriched atmosphere. J Pest Sci. 82:7–11.
- Liu QN, Bian DD, Jiang SH, Li ZX, Ge BM, Xuan FJ, Yang L, Li FC, Zhang DZ, Zhou CL, Tang BP. 2014. The complete mitochondrial genome of the red flour beetle, Tribolium castaneum (Coleoptera: Tenebrionidae). Mitochondrial DNA. 10:1–3.
- Nan XN, Wei C, He H. 2016. The complete mitogenome of Eucryptorrhynchus brandti (Harold) (Insecta: Coleoptera: Curculionidae). Mitochondrial DNA. 27:2060–2061.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.
- Yuan ML, Zhang QL, Zhang L, Guo ZL, Liu YJ, Shen YY, Shao RF. 2016. High-level phylogeny of the Coleoptera inferred with mitochondrial genome sequences. Mol Phylogenet Evol. 104:99–111.
- Zhang ZQ, Wang XJ, Li RZ, Guo RJ, Zhang W, Wang S, Hao CF, Wang HP, Li ML. 2014. The mitochondrial genome of Dastarcus helophoroides (Coleoptera: Bothrideridae) and related phylogenetic analyses. Gene. 560:15–24.
