Abstract
The complete mitochondrial genome of the hybrid grouper Epinephelus coioides ♀ × Epinephelus akaara ♂ is presented in this study. The mitochondrial genome is 16,458bp long and consists of 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes and a control region. The gene order and composition of the hybrid grouper mitochondrial genome was similar to that of most other vertebrates. The nucleotide compositions of the light strand in descending order is 28.87% of G, 28.26% of T, 26.58% of A and 15.79% of C. With the exception of the NADH dehydrogenase subunit 6 (ND6) and eight tRNA genes, all other mitochondrial genes are encoded on the heavy strand. The phylogenetic analysis by maximum-likelihood (ML) method showed that the hybrid grouper has the closer relationship to the hybrid grouper Epinephelus coioides ♀ × Epinephelus lanceolatus ♂.
Epinephelus coioides and Epinephelus akaara, both belonged to Serranidae in the order Perciformes. Epinephelus coioides, named as the orange-spotted grouper, serves as an important commercial mariculture fish species, due to its excellent food quality, abundant nutrients and rapid growth, which has been widely distributed in South China and Southeast Asian countries (Lin et al. Citation2016). Epinephelus akaara, commonly named as the Hong Kong grouper, is widely distributed in Northwest Pacific including southern China, Taiwan, East China Sea, Korea, and southern Japan (Xie et al. Citation2016). The hybrid grouper (Epinephelus coioides ♀ × Epinephelus akaara ♂) is inclined to take Epinephelus coioides as female parent, with grouper Epinephelus akaara as male parent of hybrid F1 generation. As the hybrid generation has a high potential economic value, and there is little information about its genetic characteristics, we use the next-generation sequencing techniques to study the complete mitochondrial genome of the hybrid grouper (Epinephelus coioides ♀ × Epinephelus akaara ♂), in order to develop new DNA markers for the studies on population genetics of the hybrid grouper (Xie et al. Citation2015). The specimen was obtained from Daya Bay Fishery Development Center, Guangdong, China. The total genomic DNA was extracted from the fin of the fresh fish using the salting-out procedure (Howe et al. Citation1997).
The complete mitochondrial genome of the hybrid grouper Epinephelus coioides ♀ × Epinephelus akaara ♂ (Genbank accession number KX575834) is 16,458 bp in length, consisting of 13 protein-coding genes, 2 ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA genes (tRNA) and 1 control region (), which is the same as the typical vertebrates (Wang et al. Citation2008). Most of the genes are encoded on the heavy strand, with only the NADH dehydrogenase subunit 6 (ND6) and eight tRNA genes [Gln, Ala, Asn, Cys, Try, Glu, Pro, Ser (TGA)] encoded on the light strand. Overall nucleotide compositions of the light strand are 26.58% of A, 15.79% of C, 28.26% of T and 28.87% of G. However, the most representative base is G and the bias against C was observed, which is different from the base compositions of mitochondrial genome of other teleosts.
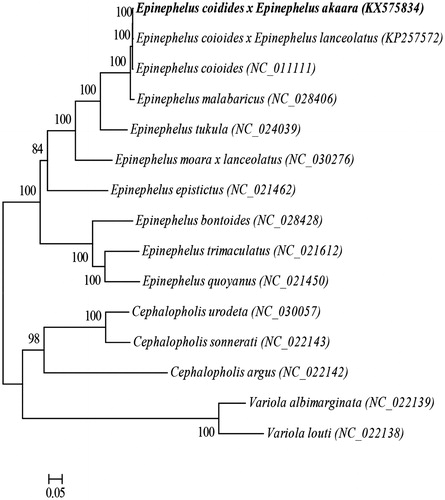
Figure 1. The ML phylogenetic tree of Perciformes species. Numbers on each node are bootstrap values of 100 replicates.
All the protein-coding genes begin with an ATG start codon except for COX1 started with GTG and ATP6 started with CTG. Four types of top codons revealed are TAA (COX1, ATP8, ATP6, ND4L, ND5, ND1), TA(COX3), T (ND2, COX2, ND3, ND4, CYTB) and TAG (ND6). The 12S and 16S rRNA genes are located between the tRNA-Phe (GAA) and tRNA-Leu (TAA) genes, and are separated by the tRNA-Val gene with the same situation found in other vertebrates. Most genes are either abutted or overlapped. The 22 tRNA genes vary from 68 to 76 bp in length. All these could be folded into the typical cloverleaf secondary structure although numerous non-complementary and T–G base pairs exist in the stem regions. The control region was 760 bp in length, located between tRNA- Pro (TGG) and tRNA-Phe (GAA) gene.
The phylogenetic position of the hybrid grouper Epinehelus_moara ♀ × Epinephelus lanceolatus ♂ was reconstructed with the complete mtDNA sequences from 15 species of Perciformes by using the maximum-likelihood (ML) methods (Kumar et al. Citation2004). As shown in , the hybrid grouper Epinehelus_moara ♀ ×Epinephelus lanceolatus ♂ has the closer relationship to the hybrid grouper Epinephelus coioides ♀ ×Epinephelus lanceolatus ♂.
Disclosure statement
The authors report no declaration of interest. The authors alone are responsible for the content and writing of this article.
Funding
This work is supported by the Guangdong Provincial Natural Science Foundation (2015A030313069, 2015A030313409), the Guangdong Provincial Science and Technology Program (2013B020308001, 2014B090905008), Pearl River S&T Nova Program of Guangzhou (2013J2200093). The Special Fund for Fisheries-Scientific Research of Guangdong Province (A201400A01, A201501A03, A201501A09) and the Fundamental Research Funds for the Central Universities (161gzd14, 161gpy35)
References
- Howe JR, Klimstra DS, Cordon-Cardo C. 1997. DNA extraction from paraffin-embedded tissues using a salting-out procedure: a reliable method for PCR amplification of archival material. Histol Histopathol. 12:595–601.
- Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150–163.
- Lin K, Zhu Z, Ge H, Zheng L, Huang Z, Wu S. 2016. Immunity to nervous necrosis virus infections of orange-spotted grouper (Epinephelus coioides) by vaccination with virus-like particles. Fish Shellfish Immunol. 56:136–143.
- Xie Z, Li S, Yao M, Lu D, Li Z, Zhang Y, Lin H. 2015. The complete mitochondrial genome of the Trachinotus ovatus (Teleostei, Carangidae). Mitochondrial DNA. 26:644–646.
- Xie Z, Xiao L, Wang X, Tang L, Liu Y, Chen H, Li S, Zhang Y, Lin H. 2016. The complete mitochondrial genome of the Epinephelus akaara (Perciformes: Serranidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1890–1891.
- Wang C, Chen Q, Lu G, Xu J, Yang Q, Li S. 2008. Complete mitochondrial genome of the grass carp (Ctenophar yngodon idella, Teleostei): insight into its phylogenic position within Cyprinidae. Gene. 424:96–101.
