Abstract
The genus Discocheilus is endemic to China and only distributed in the Pearl River. The complete mitochondrial genome of Discocheilus wui and Discocheilus wuluoheensis were successfully sequenced in this study. The complete mitochondrial genomes of these two species are 16,589 bp and 16,587 bp in length, respectively. They both consist of 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes and a control region. The gene compositions and genome organizations of the two species are similar to other fish. The complete mitochondrial genome sequence of these two species will contribute to reveal the genetic backgrounds for their conservation.
The genus Discocheilus is a member of subfamily Labeoninae in the family Cyprinidae and is endemic to the Pearl River in China (Li et al. Citation1996; Zhang et al. Citation2000). Up to now, only a few studies involved the phylogenetic positions of Discocheilus wui and Discocheilus wuluoheensis have been reported (Zheng et al. Citation2010; Citation2016). The complete mitochondrial genomes of the two species were presented in this study. The specimens sequenced in this study are deposited in the Kunming Institute of Zoology, Chinese Academy of Sciences. The sequences were deposited in GenBank with the accession numbers KX840358 and KX840359, respectively. Discocheilus wui (voucher no. KIZ20080045) was collected from Leye, Guangxi (24°40′03.6″N, 106°25′13.5″E); and Discocheilus wuluoheensis (voucher no. KIZ20080429) was collected from Luoping, Yunnan (24°45′49.1″N, 104°29′46.0″E).
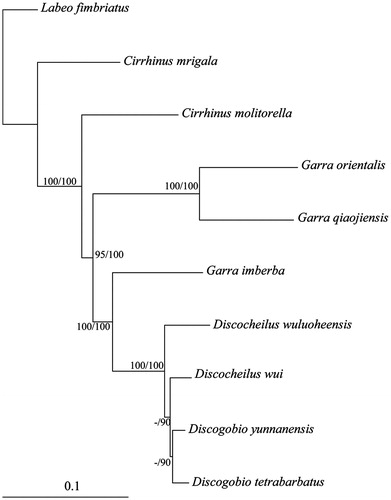
Total DNA was extracted from the fin using the standard phenol-chloroform method, and pair-end sequencing were generated from Illumina Hiseq 2500. The DNA sequences were annotated using MitoAnnotator (Iwasaki et al. Citation2013), and nucleotide composition were calculated by MEGA7 (Kumar et al. Citation2016). The gene compositions and genome organizations of the two species are similar to other fish (Miya et al. Citation2003). The complete mitochondrial genome of D. wui and D. wuluoheensis are 16,589 and 16,587 bp in length, respectively. They both include 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and a control region (D-loop). The overall base composition of the heavy strand are: A 32.2%, T 26.6%, C 26.0%, G 15.3% for D. wui, and A 32.1%, T 26.6%, C 25.9%, G 15.4% for D. wuluoheensis. Both AT contents are higher than GC contents, which are consistent with other fishes (Miya et al. Citation2003). All genes of these two species are encoded on the H-strand, with the exception of one protein-coding gene (ND6) and eight tRNA genes. Two overlaps were identified between protein-coding genes (ATP8–ATP6, and ND4L–ND4) on the same strand. Although ND5 and ND6 also overlapped, they were encoded on the opposite strands. All protein-coding genes start with an ATG codon except for COI with a GTG start codon. Six genes end with complete stop codon (TAA) while seven genes have incomplete stop codon (TA or T), which were presumably completed as TAA by post-transcriptional polyadenylation (Nardi et al. Citation2001). All tRNA genes of two species ranged from 66 bp to 76 bp and 67 bp to 76 bp in length respectively. The 12S and 16S rRNA genes were 952 bp and 1684 bp in length for D. wui, while 952 bp and 1685 bp for D. wuluoheensis respectively. The control regions are 934 bp and 931 bp in length, respectively. The phylogenetic tree showed that D. wui and D. wuluoheensis are most closely related to the genus Discogobio (), and the results are consistent with that of several mitochondrial genes (Zheng et al. Citation2010). The data of mitochondrial genome combined with the nuclear genes will be better to understand the genetic relationships of these endemic fish. The complete mitochondrial genome sequence of these two species will contribute to reveal the genetic backgrounds for their conservation.
Figure 1. Molecular phylogeny of Discocheilus and the related species in Labeoninae based on complete mitochondrial genome. The phylogenic tree is constructed with maximum likelihood and Bayesian method. The nodal numbers are ML bootstrap and BPP values, respectively. Only values above 50% are given. The accession number for tree construction is listed as follows: Labeo fimbriatus (KP025676), Cirrhinus mrigala (JQ838173), Cirrhinus molitorella (KF160921), Garra imberba (KM255666), Discogobio yunnanensis (KJ997760), Discogobio tetrabarbatus (KJ669372), Garra orientalis (JX290078), and Garra qiaojiensis (KF727438).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding
This work was supported by the National Natural Science Foundation of China (31201707), and CAS “Light of West China” Program.
References
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh T, Sado T, Mabuchi K, Takeshima H, Miya M, Nishida M. 2013. Mitofish and mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Li WX, Lu ZM, Mao WN, Sun RF. 1996. A new species of Discolabeo from Yunnan. Chinese J Fish. 9:20–22.
- Miya M, Takashima H, Endo H, Ishiguro NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, et al. 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol. 26:121–138.
- Nardi F, Carapelli A, Fanciulli PP, Dallai R, Frati F. 2001. The complete mitochondrial DNA sequence of the basal hexapod Tetrodontophora bielanensis: evidence for heteroplasmy and tRNA translocations. Mol Biol Evol. 18:1293–1304.
- Zhang E, Yue PQ, Chen JX. 2000. Labeoninae. In: Yue PQ, editor. Fauna sinica Osteichthyes Cypriniformes III.Beijing: Science Press. p. 268–272.
- Zheng LP, Chen XY, Yang JX. 2016. Molecular systematics of the Labeonini inhabiting the karst regions in southwest China (Teleostei, Cypriniformes). ZooKeys. 612:133–148.
- Zheng LP, Yang JX, Chen XY, Wang WY. 2010. Phylogenetic relationships of the Chinese Labeoninae (Teleostei, Cypriniformes) derived from two nuclear and three mitochondrial genes. Zool Scr. 39:559–571.
