Abstract
We characterized the complete mitogenome sequence of the Cape honey bee, Apis mellifera capensis, from South Africa. The circle genome is 16,470 bp in length, with the base composition of 43.2% A, 9.6% C, 5.6% G, and 41.5% T. The assembled mitogenome has 13 protein-coding genes (PCGs), 22 transfer RNAs, two ribosomal RNA genes, and one control region. All protein-coding genes are initiated by ATT, ATC, ATG or ATA codons and are terminated by the typical stop codon TAA. The heavy strand encodes four protein-coding genes, eight tRNAs, and two rRNAs. The light strand encodes nine protein-coding genes and 14 tRNAs. The complete mitogenome sequence of A.m. capensis is identical to the gene arrangement found in other A. mellifera mitogenomes and it provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis of members of the genus Apis.
Africa is the home to at least 10 indigenous Apis mellifera L. (western honey bee) subspecies that are distributed across the continent with substantial geographical, climatic, and ecological variability (Gupta et al. Citation2014). Apis mellifera scutellata and Apis mellifera capensis are two important subspecies inhabiting South Africa (Hepburn & Radloff Citation1998). Apis mellifera capensis, the Cape honey bee, is a facultative social parasite (Cape bee workers invade other non-Cape bee colonies, becoming the resident reproductive), reproducing thelytokously (unfertilized eggs become diploid females), and is characterized by a unique set of genetic, behavioural, and physiological traits possessed by the worker bees (Onions Citation1912; Hepburn & Crewe Citation1991; Neumann & Moritz Citation2002).
In this study, we report the complete mitogenome (mitochondrial genome) of an A.m. capensis worker (Accession no. KX870183) collected from an apiary close to Knysna, a city that lies in the natural distribution of A.m. capensis in South Africa (34°05′S–22°99′E (Hepburn & Radloff Citation1998). The identity of the worker bee was confirmed using classic morphometrics as per Hepburn and Radloff (Citation1998). Total genomic DNA was isolated from a worker honey bee as per Hunt & Page (Citation1995) using cetyltrimethylammonium bromide (CTAB), followed by phenol:chloroform:isoamyl alcohol (25:24:1). The genomic DNA was quantified using a Qubit® 3.0 Fluorometer (Thermo Scientific Inc., Waltham, MA, USA). Genomic libraries were constructed and genome skimming (Straub et al. Citation2012) was performed using pair-end sequencing (2 × 100 bp) on the Illumina HiSeq 2000 (San Diego, California, USA) sequencing platform.
The resulting FASTQ reads were trimmed and mapped to the reference mitogenome of A.m. ligustica (L06178.1, the Italian honey bee) using Geneious R9.1 (Kearse et al. Citation2012). We used 10 iterations of custom-settings to ensure high-stringency mapping. In regions with ambiguities, we selected short consensus segments (>100 bp) and remapped these with original reads to elongate them without bias of the original reference sequence. We combined these assemblies using 2–3 iterations to achieve a final sequence that was examined by eye. To confirm, a final mapping of the reads was done to the final sequence to ensure there were no errors.
The mitochondrial genome of A.m. capensis constitutes a DNA circular closed loop that is 16,470 bp in length. The A.m. capensis mitogenome was similar in content and organization to that of A.m. ligustica. It contained 13 protein-coding genes, 22 putative tRNAs, two rRNAs, and an AT-rich control region. There was a strong A + T bias (84.7% of the genome). The heavy strand encodes four protein-coding genes, eight tRNAs, and two rRNAs. The light strand encodes nine protein-coding genes and 14 tRNAs. Six protein-coding genes of the A.m. capensis mitochondrial genome started with ATT, four with ATG, two with ATA, and one with ATC. The stop codon of each of these protein-coding genes was TAA.
The mitogenome of A.m. capensis shared 96% genetic identity with that of A.m. ligustica, with 274 nucleotide substitutions and 338 indel differences. There were 32 amino acid differences, occurring in all genes except ND4L. The starting position of ND4 differs among Apis mitogenomes. Based on the A.m. ligustica mitogenome, which has the longest ND4 annotated for an Apis mellifera mitogenome to date (411 amino acids, relative to 409 to 398 in other taxa presented in the ), the likely homologous start position in A.m. capensis resulted in a stop codon at the eighth amino acid position. However, the starting position for ND4 based on annotations of A.m. scutellata and A.m. syriaca, which exclude these initial sites, did not lead to a stop codon on the translated ND4 sequence of A.m. capensis. Thus, we assumed this was the appropriate start codon.
Figure 1. Phylogenetic relationships within the genus Apis based on maximum likelihood approach of the mitochondrial genome nucleotide sequence of the 13 protein-coding genes. The bootstrap values are given in numbers on the left of each node. The letter/number combination on the right side of each node indicates the GenBank accession numbers.
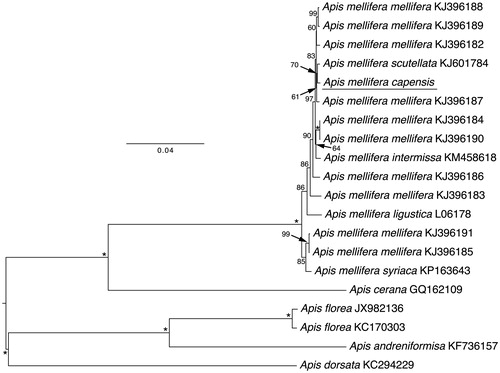
There was 99% identity of the A.m. capensis mitogenome with that of A.m. scutellata (KJ601784), a honey bee subspecies which is distributed in South Africa borders with that of A.m. capensis. The differences were due to 44 nucleotide substitutions and 95 sites with indels which were all located in non-coding regions. There were three amino acid differences between the A.m. capensis and A.m. scutellata mitogenomes, one each in ND2, CO3, and ND5. Furthermore, A.m. capensis has an additional amino acid in the ND4 gene that is absent in A.m. scutellata (but is present in the other mitogenomes). In total, there were four amino acid differences between A.m. capensis and A.m. scutellata (3 substitutions and one indel).
We also identified a pseudogene of ∼2000 bp (the exact boundaries were difficult to establish due to assembly issues). This covered ATP6 through ND5, with two large (> 500 bp) deletions, as well as many smaller insertions and deletions relative to the reference genome. Overall identification for the regions covered was over 78%, with high identity in some regions.
A phylogenetic analysis was constructed using the 13 mitochondrial protein-coding genes and two rRNAs with inclusion of 19 published mitogenomes from other species of Apis and subspecies of A. mellifera. To identify the phylogenetic position of A.m. capensis, we used RAxML 7.2.8 (Stamatakis Citation2006) and the GTRGAMMA model to construct a maximum-likelihood tree; we estimated the ML tree using 10 random additions, and assessed support with 1000 bootstrap replicates (). We used midpoint rooting. The tree confirms that A.m. capensis is within the Apis mellifera clade and is supported as sister taxa to A.m. scutellata with 70% support (). Apis mellifera capensis clusters inside A.m. mellifera subspecies, which do not form a monophyletic clade with the mitochondrial genome. The genetic distance between the A.m. capensis and A.m. scutellata mitochondrial genomes was 0.002. In contrast, the distance between the A.m. capensis genome and all other A. mellifera mitochondrial genomes averaged 0.006, consistent with genetic distances between African (0.004) and European and Middle Eastern A. mellifera (0.014) (Crozier & Crozier Citation1993; Gibson & Hunt Citation2015; Haddad Citation2015; Hu et al. Citation2015). In conclusion, the complete A.m. capensis mitogenome will provide essential and fundamental information to elucidate population genetics, biogeography and evolutionary history of this bee and related subspecies.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding
This project was financed through a cooperative agreement provided by the United States Department of Agriculture, Animal and Plant Health Inspection Service (USDA-APHIS) under the grant number 14-8130-0414-CA.
References
- Crozier RH, Crozier YC. 1993. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics. 133:97–117.
- Gibson JD, Hunt GJ. 2015. The complete mitochondrial genome of the invasive Africanized Honey Bee, Apis mellifera scutellata (Insecta: Hymenoptera: Apidae). Mitochondrial DNA. 27:561–562.
- Gupta RK, Reybroeck W, van Veen JW, Gupta A. 2014. Beekeeping for poverty alleviation and livelihood security. Vol. 1: Technological aspects of beekeeping. 1st ed. Netherlands: Springer. p. 665.
- Haddad NJ. 2015. Mitochondrial genome of the Levant Region honeybee, Apis mellifera syriaca (Hymenoptera: Apidae). Mitochondrial DNA. 30:1–2.
- Hepburn HR, Radloff SE. 1998. Honeybees of Africa. Berlin, Heidelberg, New York: Springer Verlag. p. 370.
- Hepburn HR, Crewe RM. 1991. Portrait of the Cape honeybee, Apis mellifera capensis. Apidologie. 22:567–580.
- Hunt GJ, Page RE. 1995. Linkage map of the honey bee, Apis mellifera, based on RAPD markers. Genetics. 139:1371–1382.
- Hu P, Lu ZX, Haddad N, Noureddine A, Loucif-Ayad Q, Qang YZ, Zhao RB, Zhang AL, Guan X, Zhang HX, Niu H. 2015. Complete mitochondrial genome of the Algerian honeybee, Apis mellifera intermissa (Hymenoptera: Apidae). Mitochondrial DNA. 26:1–2.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Neumann P, Moritz RFA. 2002. The Cape honeybee phenomenon: the sympatric evolution of a social parasite in real time? Behav Ecol Sociobiol. 52:271–281.
- Onions GW. 1912. South African ‘fertile worker bees’. Agric J Union S Afr. 1:720–728.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Straub SCK, Parks M, Weitemier K, Fishbein M, Cronn R, Liston A. 2012. Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am J Bot. 99:349–364.
