Abstract
The complete mitochondrial genome of the Tamarisk jird, Meriones tamariscinus, was sequenced. The 16,389bp genome contains 37 genes, typical for rodent mitogenomes, including 22 tRNA genes, 2 rRNA genes, and 13 protein-coding genes. The total GC content of the mitochondrial genome is 36.8%, with a base composition of 34.0% A, 24.5% C, 12.3% G, and 29.2% T. The phylogenetic analysis showed that M. tamariscinus was classified in the genus Meriones, Muridae.
The Tamarisk jird (Meriones tamariscinus) is a rodent that inhabits desert and semi-desert areas. The animal was collected from Khorgos (44°7′39″N, 80°24′13E), China, and was identified by morphology and DNA sequence (Gu et al. Citation2011). The complete mitochondrial genome of M. tamariscinus (GenBank accession no. KX688104) was sequenced by using long PCR and Sanger sequencing. The circular mitochondrial genome was 13,689 bp long and contained 37 genes (2 rRNA genes, 22 tRNA genes, and 13 protein-coding genes) that are typically found in rodent mitochondrial genomes () (Jiang et al. Citation2012). The base composition was 29.2% T, 24.5% C, 34.9% A and 12.3% G. The overall disproportional A + T content was 63.2%, which was higher than any of the three previously studied Meriones species (M. unguiculatus, 63%; M. libycus, 61.6%; and M. meridianus, 62.7%)(Luo & Liao Citation2016a, Citation2016b). ATG was used as the start codon of eight protein-coding genes, while ATA was used by ND1, GTG was used by ND4L, and ATT was used by ND2, ND3, and ND5. The unusual start codons ATA and ATT have also been observed in other mammals (Jiang et al. Citation2012). The coding regions of the genes Cox1, Cox2, ATPase8, ATPase6, ND3, ND4L, and ND5 were terminated by TAA, while ND6 was terminated by TAG. Five incomplete stop codons T(––) were found in the ND1, ND2, Cox3, ND4, and cytb genes. Incomplete stop codons have been observed in many other mammals (Peng et al. Citation2007). Several pairs of genes overlapped: ATPase8 and ATPase6, ATPase6 and Cox3, and ND4L and ND4 (). Two rRNA genes (12S rRNA and 16S rRNA) were identified via DOGMA with default settings and refined through alignment to homologous genes in other mitochondrial genomes. The 12S rRNA gene was 951 bp in length, and the 16S rRNA gene was 1572 bp in length. These genes had 61.5% and 63.8% A + T content, respectively. They were flanked by tRNAPhe and tRNALeu and separated by tRNAVal. The length of M. tamariscinus tRNA genes ranged from 59 to 75 bp. Of the 22 tRNA genes, 21 could fold into a cloverleaf secondary structure. The tRNAser(AGN) was only 59 bp in length and was missing the dihydrouridine stem and loop. This loss is common among vertebrates (Zhong et al. Citation2010). The putative origin of replication for the light strand (OL) is located in the WANCY cluster between tRNAAsn and tRNACys. The concatenated nucleotide sequence of the 12 protein-coding genes encoded on the H-strand from 26 available mitochondrial genomes and M. tamariscinus mitogenome were used to reconstruct the phylogenetic relationships using neighbour-joining (NJ), maximum-likelihood (ML), and minimum evolution(ME) methods (Ito et al. Citation2010). The phylogenetic methods produced the same tree topology in the branching patterns. The monophyly of the Meriones clade received relatively strong support (; BP = 100%). The phylogenetic analysis showed Rattus forming a clade with Mus, and this clade was closer to the Meriones clade than was the other clade. This finding demonstrates that Meriones, including M. libycus and M. meridianus, should be placed in Muridae.
Figure 1. A maximum-likelihood tree of M. tamariscinus and 26 rodents inferred from 12 mitochondrial genes in H-strand. Percentage bootstrap values are shown on interior branches with 1000 replicates for the ML method. GenBank accession numbers are indicated in brackets.
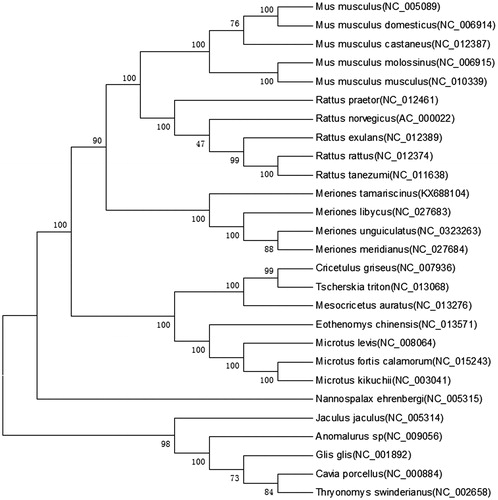
Table 1. Elements and base composition of M. tamariscinus mitochondrial genome.
Disclosure statement
The authors declare that there are no conflicts of interest.
Additional information
Funding
References
- Gu D, Zhou L, Ma Y, Ning S, Hou Y, Zhang B. 2011. Geographic variation in mitochondrial DNA sequences and subspecies divergence of the Tamarisk Gerbil (Meriones tamariscinus) in China. Acta Theriol Sin. 31:347–357. (Article in Chinese)
- Ito M, Jiang W, Sato JJ, Zhen Q, Jiao W, Goto K, Sato H, Ishiwata K, Oku Y, Chai JJ, Kamiya H. 2010. Molecular phylogeny of the subfamily Gerbillinae (Muridae, Rodentia) with emphasis on species living in the Xinjiang-Uygur Autonomous Region of China and based on the mitochondrial Cytochrome b and Cytochrome c Oxidase subunit II genes. Zool Sci. 27:269–278.
- Jiang X, Gao J, Ni L, Hu J, Li K, Sun F, Xie J, Bo X, Gao C, Xiao J, Zhou Y. 2012. The complete mitochondrial genome of Microtus fortis calamorum (Arvicolinae, Rodentia) and its phylogenetic analysis. Gene. 498:288–295.
- Luo G, Liao J. 2016a. The complete mitochondrial genome of Meriones libycus (Rodentia: Cricetidae) and its phylogenetic analysis. Mitochondrial DNA A DNA Mapp Seq Anal. 27:2545–2546.
- Luo G, Liao J. 2016b. The complete mitochondrial genome of Meriones meridianus (Rodentia: Cricetidae) and its phylogenetic analysis. Mitochondrial DNA A DNA Mapp Seq Anal. 27:2547–2548.
- Peng R, Zeng B, Meng XX, Yue BS, Zhang ZH, Zou FD. 2007. The complete mitochondrial genome and phylogenetic analysis of the giant panda (Ailuropoda melanoleuca). Gene. 397:76–83.
- Zhong HM, Zhang HH, Sha WL, Zhang CD, Chen YC. 2010. Complete mitochondrial genome of the red fox (Vuples vuples) and phylogenetic analysis with other canid species. Zool Res. 31:122–130.
