Abstract
The mitochondrial genome (mitogenome) of Eurytoma sp. (Hymenoptera: Eurytomidae: Eurytominae) was determined in this study. The sequenced region is 14,946 bp with an A + T content of 82.65%, including 13 protein-coding genes, 2 ribosomal RNAs, 18 transfer RNAs, and a partial sequence of AT-rich region. All PCGs begin with typical ATN codons and use standard canonical TAA as their termination codons, except for atp6 gene, which stops with an incomplete codon T. A comparison of all the sequenced hymenopteran mitogenomes revealed that Eurytoma sp. has dramatic gene rearrangements, some of which is peculiar to Chalcidoidea. What’s more, Maximum likelihood (ML) analysis highly supported the monophyly of both Chalcidoidea and Apocrita.
Eurytomidae is found throughout the world and over 1400 species in 84 genera are described (Lotfalizadeh et al. Citation2007; Gates Citation2008). However, no complete or nearly complete mitogenome of any species in this family has been reported. Here, we present the mitogenome of Eurytoma sp. (Hymenoptera: Eurytomidae), which was collected in Taiyuan City, Shanxi Province, China (37° 52′ 24.62″ N, 112° 32′ 51.17″ E). Now the specimen is stored in Institute of Zoology, Chinese Academy of Sciences (accession number: 20100812A).
The nearly complete mitogenome of Eurytoma sp. is 14,946 bp in length with the A + T content of 82.65%, containing 13 protein-coding genes (PCGs), 2 ribosomal RNAs (rrnL and rrnS), 18 transfer RNAs (tRNAs), and a partial AT-rich region. Analysis of the sequenced region revealed dramatic mitochondrial gene rearrangements, including not only tRNAs, but also seven protein-coding genes, which are similar to some other species of Chalcidoidea (Oliveira et al. Citation2008; Xiao et al. Citation2011). Additionally, 13 intergenic spacers (154 bp in total) and 13 overlapping regions (39 bp in total) are dispersed throughout the genome.
All 13 PCGs are initiated by typical ATN codons (three ATA, four ATT, and six ATG) and stop with standard termination codon TAA, except for atp6, which ends with a single T. In total, 18 of the 22 tRNA genes were sequenced. All the available tRNAs share typical cloverleaf structures except for trnS2, whose dihydrouridine (DHU) arm forms a simple loop, as seen in other hymenopterans (Wei et al. Citation2010; Huang et al. Citation2014; Zhang et al. Citation2015). The rrnL and rrnS genes are 1290 bp and 747 bp, with their A + T content of 87.05% and 86.33%, respectively. Both of them are located on the minority stand and separated by trnA and trnQ, which is unique in the available hymenopterans.
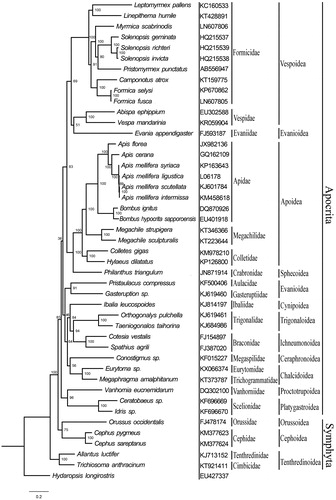
We conducted a phylogenetic analysis using all the complete hymenopteran mitogenomes that were available in GenBank, along with a Hemipteran outgroup species (Hydaropsis longirostris). The phylogenetic relationships based on nucleotide sequences of 13 PCGs using Maximum likelihood (ML) method revealed the monophyly of Apocrita (). Within the Apocrita, all of the superfamilies, except for Evanioidea, were proved as monophyletic groups. The results were similar to some other studies about the phylogeny and classification of Hymenoptera (Dowton & Austin Citation1994; Sharkey Citation2007; Huang et al Citation2016). Although there were only two species of Chalcidoidea, the monophyly was also highly supported ().
Figure 1. Maximum likelihood tree of Hymenoptera, inferred from nucleotide sequences of 13 mitochondrial PCGs. The tree was rooted using the outgroup taxon Hydaropsis longirostris (Hemiptera). Each species involved in the tree has a Genbank accession number on the right side of its scientific name. Numbers denote bootstrap values in percentages.
Nucleotide sequence accession number
The mitogenome sequence of Eurytoma sp. has been assigned GenBank accession number KX066374.
Disclosure statement
The authors alone are responsible for the content and writing of the paper, and report no conflicts of interest.
Funding
This work was supported by the National Natural Science Foundation of China, 10.13039/501100001809 (grant nos. 31372249, 31572298) and the Cooperative Research Projects in Biodiversity jointly supported by the National Natural Science Foundation of China and the K.T. Li Foundation for the Development of Science and Technology (grant no.31561163003), awarded to APL.
References
- Dowton M, Austin AD. 1994. Molecular phylogeny of the insect order Hymenoptera: apocritan relationships. Proc Natl Acad Sci USA. 91:9911–9915.
- Gates M. 2008. Species revision and generic systematics of world Rileyinae. Berkeley and Los Angeles, CA: University of California Press Publications in Entomology; p. 126–128.
- Huang DY, Su TJ, He B, Gu P, Liang AP, Zhu CD. 2016. Sequencing and characterization of the Megachile strupigera (Hymenoptera: Megachilidae) mitochondrial genome. Mitochondrial DNA Part B. 1:309–311.
- Huang DY, Su TJ, Qu L, Wu YP, Gu P, He B, Xu XF, Zhu CD. 2014. The complete mitochondrial genome of the Colletes gigas (Hymenoptera: Colletidae: Colletinae). Mitochondrial DNA. doi: 10.3109/19401736.2014.987243.
- Lotfalizadeh H, Delvare G, Rasplus JY. 2007. Phylogenetic analysis of Eurytominae (Chalcidoidea: Eurytomidae) based on morphological characters. Zool J Linn Soc. 151:441–510.
- Oliveira DCSG, Raychoudhury R, Lavrov DV, Werren JH. 2008. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (Hymenoptera: Pteromalidae). Mol Biol Evol. 25:2167–2180.
- Sharkey MJ. 2007. Phylogeny and classification of Hymenoptera. Zootaxa. 1668:521–548.
- Wei SJ, Tang P, Zheng LH, Shi M, Chen XX. 2010. The complete mitochondrial genome of Evania appendigaster (Hymenoptera: Evaniidae) has low A + T content and a long intergenic spacer between atp8 and atp6. Mol Biol Rep. 37:1931–1942.
- Xiao JH, Jia JG, Murphy RW, Huang DW. 2011. Rapid evolution of the mitochondrial genome in Chalcidoid wasps (Hymenoptera: Chalcidoidea) driven by parasitic lifestyles. PLoS One. 6:e26645.
- Zhang Y, Su TJ, He B, Gu P, Huang DY, Zhu CD. 2015. Sequencing and characterization of the Megachile sculpturalis (Hymenoptera: Megachilidae) mitochondrial genome. Mitochondrial DNA. doi: 10.3109/19401736.2015.1122774.
