Abstract
We present the complete mitochondrial genome sequence of Plakobranchus cf. ocellatus (Heterobranchia: Sacoglossa), a so-called ‘solar-powered’ sea slug with long-term retention of chloroplasts. The mitochondrial genome was 14,177 bp in length containing the standard set of 13 protein-coding genes, 2 rRNAs, and 22 tRNAs. The base composition of 27.3% A, 15.6% C, 18.6% G, and 38.5% T showed a strong A + T bias. The genome organization of P. cf. ocellatus is identical to the other sacoglossan mitogenomes sequenced so far, except for Ascobulla fragilis.
Plakobranchus cf. ocellatus van Hasselt, 1824 belongs to a small group of marine, heterobranch slugs, called Sacoglossa, of which certain species have the ability to sequester chloroplasts from its food algae (Hirose Citation2005; Christa et al. Citation2012). These ‘stolen’ plastids (kleptoplasts) are then stored in a functional state in the digestive gland cells of the slugs and allow them to endure weeks (short-term retention) or months (long-term retention) of starvation during which time the kleptoplasts continue photosynthesis inside the slugs (Trench et al. Citation1970; Christa et al. Citation2012, Citation2014; Wägele & Martin Citation2014; Vries et al. Citation2015). Here, we present the complete mitochondrial genome of P. cf. ocellatus, a so-called ‘solar-powered’ sea slug with long-term retention (Händeler et al. Citation2009; Christa et al. Citation2012).
Specimens of P. cf. ocellatus were collected on the Philippines (10°14'30.4"N 124°03'47.2"E) in December 2012. The heads of about 60 slugs were dissected to remove the post-pharyngeal nerve ring, which were directly deep frozen in liquid nitrogen and then stored (−80 °C). From these, genomic DNA was extracted using the QIAGEN DNeasy® Blood & Tissue Kit (Hilden, Germany). Voucher material is stored in absolute ethanol at the Zoological Research Museum Alexander Koenig (voucher no. ZFMK-TIS-29490).
Two genomic libraries (insert sizes: 350 and 550 bp, resp.) were prepared using the TruSeq® DNA PCR-Free Library Preparation Kit (San Diego, CA) according to the manufacturer’s protocol and 125 bp paired-end reads were sequenced on an Illumina HiSeq 2500 platform (San Diego, CA). All read pairs were used for mitochondrial genome assembly with MITObim 1.8 (Hahn et al. Citation2013), using the sequence of the mitochondrial cytochrome oxidase I gene of P. ocellatus (NCBI accession number: JX272720.1) as seed. After verification of correct circularity, genome annotation was done with MITOS revision 656 (Bernt et al. Citation2013), followed by manual correction using Geneious version 7.1.9 (Kearse et al. Citation2012).
The complete mitochondrial genome (GenBank accession number: KX853083) had a length of 14,177 bp encoding for 13 protein-coding genes, 2 rRNAs, and 22 tRNAs. The base composition is 27.3% A, 15.6% C, 18.6% G, and 38.5% T. A total of 183 bp nucleotides were observed in multiple small intergenic regions, ranging from 1 to 54 bp (found between the genes coding for COX3 and trnI(gat)). The gene order was as follows: trnK(ttt), cox1, trnV(tac), rrnL, trnL1(tag), trnA(tgc), trnP(tgg), nad6, nad5, nad1, trnW(tca), trnY(gta), nad4l, cob, trnD(gtc), trnF(gaa), cox2, trnG(tcc), trnH(gtg), -trnQ(ttg), -trnL2(taa), -atp8, -trnN(gtt), trnC(gca), -atp6, -trnR(tcg), -trnE(ttc), -rrnS, -trnM(cat), -nad3, -trnS2(tga), trnS1(gct), nad4, -trnT(tgt), -cox3, trnI(gat), nad2 and is therefore identical to the other sacoglossan mitogenomes sequenced so far, i.e. Elysia chlorotica, E. ornata, Thuridilla gracilis, and Placida sp. 1 NY-2013, except for Ascobulla fragilis.
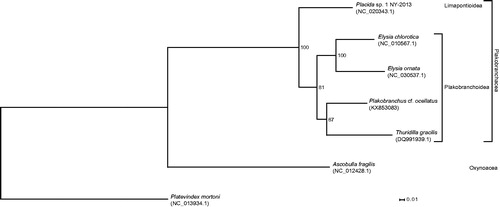
The amino acid sequence of the 13 protein-coding genes of P. cf. ocellatus and all other sacoglossan species with fully sequenced mitogenome were extracted, aligned with MAFFT v7.271 (G-INS-i mode) (Katoh & Standley Citation2013), conspicuous sites as suggested by Aliscore v1.2 (Misof & Misof Citation2009) were removed, and remaining sites concatenated into a supermatrix. Optimal model parameters and the best partitioning scheme were searched with PartitionFinder v2.0.0-pre14 (Lanfear et al. Citation2014). Phylogenetic relationships, with partitioning and optimal model parameters being considered, were inferred (with 150 bootstrap replicates as found sufficient by the MRE-based bootstopping criterion) with RAxML version 8.2.9 (Stamatakis Citation2014) ().
Figure 1. The molecular phylogeny of Plakobranchus cf. ocellatus and other sacoglossans (outgroup: Platevindex mortoni) based on the amino acid sequence of all protein-coding genes. The complete mitogenomes were downloaded from GenBank and the phylogenetic tree was constructed under the maximum-likelihood optimality criterion (150 bootstrap replicates).
Acknowledgements
We thank Margrieta Alblas, Claudia Etzbauer, Kathleen Keppler, and Sandra Kukowka for technical and laboratory support. We wish to thank Frank Richter for supplying live animals of Plakobranchus cf. ocellatus via his aquarium shop in Chemnitz, Germany.
Disclosure statement
The authors declare no conflicts of interest. All authors are responsible for the content and writing of this article.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Christa G, Wescott L, Schäberle TF, König GM, Wägele H. 2012. What remains after 2 months of starvation? Analysis of sequestered algae in a photosynthetic slug, Plakobranchus ocellatus (Sacoglossa, Opisthobranchia), by barcoding. Planta. 237:559–572.
- Christa G, Zimorski V, Woehle C, Tielens AGM, Wägele H, Martin WF, Gould SB. 2014. Plastid-bearing sea slugs fix CO2 in the light but do not require photosynthesis to survive. Proc R Soc B. 281:20132493.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129.
- Händeler K, Grzymbowski YP, Krug PJ, Wägele H. 2009. Functional chloroplasts in metazoan cells – a unique evolutionary strategy in animal life. Front Zool. 6:28.
- Hirose E. 2005. Digestive system of the sacoglossan Plakobranchus ocellatus (Gastropoda: Opisthobranchia): light- and electron-microscopic observations with remarks on chloroplast retention. Zoolog Sci. 22:905–916.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A. 2014. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol Biol. 14:82.
- Misof B, Misof K. 2009. A Monte Carlo approach successfully identifies randomness in multiple sequence alignments: a more objective means of data exclusion. Syst Biol. 58:21–34.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Trench ME, Trench RK, Muscatine L. 1970. Utilization of photosynthetic products of symbiotic chloroplasts in mucus synthesis by Placobranchus ianthobapsus (Gould), opisthobranchia, sacoglossa. Comp Biochem Physiol. 37:113–117.
- Vries J, de Woehle C, Christa G, Wägele H, Tielens AGM, Jahns P, Gould SB. 2015. Comparison of sister species identifies factors underpinning plastid compatibility in green sea slugs. Proc R Soc Lond B Biol Sci. 282:20142519.
- Wägele H, Martin WF. 2014. Endosymbioses in Sacoglossan seaslugs: plastid-bearing animals that keep photosynthetic organelles without borrowing genes. In: Löffelhardt W, editor. Endosymbiosis [Internet]. Vienna: Springer Vienna; p. 291–324.
