Abstract
The Pacific cod Gadus macrocephalus is a commercially important species belonging to the family Gadidae. In this study, we performed the first sequencing and assembly of the complete mitochondrial genome of G. macrocephalus. The complete mitochondrial genome is 16,567 bp long, consisting of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a control region. It has the typical vertebrate mitochondrial gene arrangement. Phylogenetic analysis using the mitochondrial genomes of 15 species showed that G. macrocephalus clusters with G. ogac. This mitochondrial genome provides potentially important resources for performing population genetic analysis and addressing phylogenetic issues.
The Pacific cod Gadus macrocephalus is a species of teleost fish belonging to the family Gadidae. It is widely distributed in the North Pacific, from the Yellow Sea through the Okhotsk and Bering Seas to California in the eastern North Pacific Ocean (Grant et al. Citation1987). Gadus macrocephalus is one of the commercially and ecologically significant species in cold-water marine systems. However, as a consequence of the environmental effects of human activities, the quantity of wild stocks of G. macrocephalus has declined markedly in recent decades (Maschner et al. Citation2008; FAO Citation2016). In this study, we determined the complete mitochondrial genome of G. macrocephalus, and analyzed the phylogenetic relationship of this species among Gadiformes fishes.
The G. macrocephalus specimen used in this study was collected from Donghae-si, Gangwon Province, South Korea (37.34N, 129.06E). Total genomic DNA was extracted from a tissue deposited in the National Marine Biodiversity Institute of Korea (Voucher No. MABIK TV00000001). The mitogenome was amplified in its entirety using a long PCR technique (Cheng et al. Citation1994). We used fish-versatile PCR primers in various combinations to amplify contiguous, overlapping segments of the entire mitogenome. All of the experimental procedures were carried out following the previously described methods (Miya & Nishida Citation1999).
The complete mitochondrial genome of G. macrocephalus (GenBank accession no. AP017650) is 16,567 bp long, and includes 13 protein-coding genes, 22 tRNA genes, and two rRNA genes. The ND6 gene and eight tRNA genes are encoded on the light strand. The overall base composition of the heavy strand is 27.88% A, 25.86% C, 16.86% G, and 29.38% T. Similar to the mitogenomes of other vertebrates, the AT content is higher than the GC content (Saccone et al. Citation1999). All of the tRNA genes can fold into a typical cloverleaf structure, with lengths ranging from 66 to 74 bp. The 12S rRNA (953 bp) and 16S rRNA genes (1665 bp) are located between tRNAPhe and tRNAVal and between rRNAVal and tRNALeu(UUR), respectively. Of the 13 protein-coding genes, 12 start with ATG; the exception being COI, which starts with GTG. Seven of the 13 protein-coding genes were shown to terminate with incomplete stop codons, T (ND2, COII, ND3, ND4, and Cytb) and TA (ATP6 and COIII), whereas the remaining six genes ended with complete stop codons of TAG, TAA, or AGG. A control region (868 bp) is located between tRNAPro and tRNAPhe.
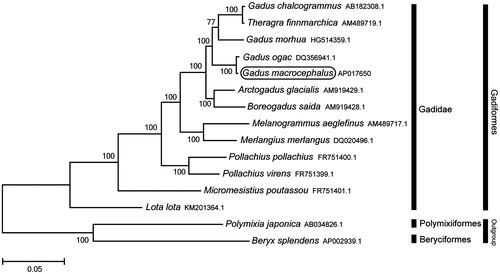
Phylogenetic trees were constructed by the maximum-likelihood method with 1000 replicates using MEGA 7.0 software (MEGA, PA, USA) (Kumar et al. Citation2016) for the newly sequenced genome and 14 further complete mitochondrial genome sequences downloaded from the National Center for Biotechnology Information. We confirmed that G. macrocephalus is clustered with G. ogac (Coulson et al. Citation2006) and rooted with the other Gadidae species (). We anticipate that the mitogenome of G. macrocephalus will provide useful information for phylogenetic and population genetic analyses.
Figure 1. Phylogenetic position of Gadus macrocephalus based on a comparison with the complete mitochondrial genome sequences of 12 Gadiformes species and the two outgroups species. The analysis was performed using the MEGA 7.0 software. The accession number of each species is indicated after the scientific name.
Disclosure statement
The authors report no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Funding
This work was supported by National Marine Biodiversity Institute Research Program (2016M00500).
References
- Cheng S, Higuchi R, Stoneking M. 1994. Complete mitochondrial genome amplification. Nat Genet. 7:350–351.
- Coulson MW, Marshall HD, Pepin P, Carr SM. 2006. Mitochondrial genomics of gadine fishes: implications for taxonomy and biogeographic origins from whole-genome data sets. Genome. 49:1115–1130.
- Food and Agriculture Organization of the United Nations (FAO). 2016. The state of world fisheries and aquaculture. Rome, Italy: Food and Agriculture Organization of the United Nations.
- Grant WS, Zhang CI, Kobayashi T, Ståhl G. 1987. Lack of genetic stock discretion in Pacific cod (Gadus macrocephalus). Can J Fish Aquat Sci. 44:490–498.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Maschner HD, Betts MW, Reedy-Maschner KL, Trites AW. 2008. A 4500-year time series of Pacific cod (Gadus macrocephalus) size and abundance: archaeology, oceanic regime shifts, and sustainable fisheries. Fish Bull. 106:386–394.
- Miya M, Nishida M. 1999. Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar Biotechnol. 1:416–426.
- Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A. 1999. Evolutionary genomics in metazoa: the mitochondrial DNA as a model system. Gene. 238:195–209.
