Abstract
In this study, the complete mitogenome of the tigertooth croaker Otolithes ruber was first determined. This mitogenome is 16,589 bp in length, and consists of 37 genes with the typical gene order and direction of transcription in vertebrates. The overall nucleotide composition is: 27.4% A; 29.1% C; 16.1% G and 27.4% T. Sizes of the 22 tRNA genes range from 66 to 74 bp. Four start codons (ACG, CTG, GTG and ATG) and three stop (AGA, TAG and TAA/TA/T) codons were detected in 13 protein-coding genes. In the Bayesian treebased on the complete mitogenomes of 18 species (including O. ruber) from the family Sciaenidae, all nodes were strongly supported. The phylogenetic results suggested that O. ruber was closed to the black-spotted croaker Protonibea diacanthus.
The family Sciaenidae (Perciformes) is commonly known as croakers and drums. As an economically important group of fishes, it comprises about 270 species in 70 genera in the world (Nelson Citation2006). The tigertooth croaker Otolithes ruber is an widespread sciaenid species inhabiting shallow coastal waters down to 40 m in the Indian and West Pacific Oceans, west to South Africa, east to southern China and Queensland, Australia (Sasaki Citation2001). The species feeds mainly on fishes and prawns with maximum total length of 90 cm (http://www.fishbase.org). In this study, we presented the complete mitochondrial genome of O. ruber and assessed its phylogenetic relationship based on another 17 available mitogenomes in the family Sciaenidae with 2 available mitogenomes in the family Epinephelidae as an outgroup.
One specimen of O. ruber (MJ2015081503) was collected by a bottom trawler in the Min River Estuary, Fujian Province, China. The protocol and data analysis methods followed Chen et al. (Citation2014). The complete mitochondrial genome of O. ruber is 16,589 bp in length (GenBank accession number: KX929060) with the typical gene order and transcriptional direction in vertebrates. It contains two rRNA genes, 22 tR 45 NA genes, 13 protein-coding genes and one control region. The overall nucleotide composition is as follows: 27.4% A, 29.1% C, 16.1% G and 27.4% T. In the 13 identified protein-coding genes, four start codons (ACG, 48 CTG, GTG and ATG) were detected; ND1 was initiated by the ACG codon, ATP8 by the GTG codon, ATP6 by the CTG codon and the other 10 genes by the ATG codon. Three stop codons (AGA, TAG and TAA/TA/A) were found; COX1 was terminated by the AGA codon, ND1 by the TAG codon and the other 11 protein-coding genes by either the TAA or incomplete T or TA codon that may form complete termination signal UAA via post-transcriptional polyadenylation (Ojala et al. Citation1981). The 12S (958 bp) and 16S (1752 bp) rRNA genes are located between the tRNA-Phe and tRNA-Leu1 genes, separated by the tRNA-Val gene. The lengths of 22 tRNA genes range from 66 to 74 bp; 21 tRNAs can be folded into the typical cloverleaf secondary structures with the exception of tRNA-Ser2 in which the DHU arm was replaced by a simple loop. A 40 bp inserted sequence was identified as the putative origin of L-strand replication (OL). The control region was 838 bp in length with high A + T (64.5%) and low G + C (35.5%) composition.
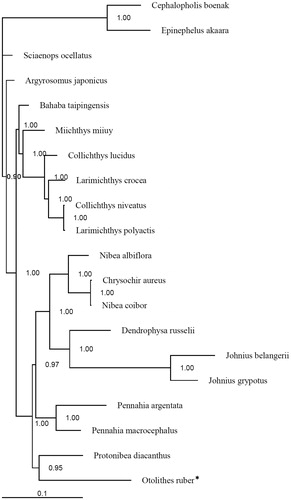
Published mitogenomes of all 18 species of the family Sciaenidae (including O. ruber in this study), and the chocolate hind Cephalopholis boenak and the Hong Kong grouper Epinephelus akaara from the family Epinephelidae were used to assess the phylogenetic relationship of O. ruber. Phylogenetic tree was constructed with the partitioned Bayesian method based the dataset combined by three partitions (the alignments of the 1, 2 codon positions of 12 H-strand encoded protein-coding genes together with 2 rRNAs) under the GRT + I + G model (Ronquist & Huelsenbeck Citation2003). As the phylogenetic tree showed, all nodes were strongly supported with high value 67 of posterior probability (). The result suggested that O. ruber was placed closely to Protonibea diacanthus, and subsequently to the group with genera Pennahia, Nibea, Chrysochir, Dendrophysa and Johnius. The resulting relationships are not consistent with the earlier conclusions based on the morphological characters. The genera Otolithes, Chrysochir, Sonorolux, Larimichthys and Collichthys formed a monophyletic group by having the distal end of the sulcus tail circular in their otolith (Sasaki Citation1989). It was also suggested that Otolithes and Chrysochir had a single synapomorphy, canines present (Sasaki Citation1989). A recent study based on mitochondrial (Cyt b, COX1) and nuclear (exon 3 of RAG1, RH, exon 2 of EGR1 and exon 1, intron 1 and exon 2 of EGR2B) genes revealed that Otolithes, Pterotolithus and Atrobucca formed a monophyletic group which closed to a large group contained Chrysochir, Protonibea, Megalonibea, Pennahia, Nibea, Dendrophysa, Austronibea, Daysciaena and Johnius (Lo et al. Citation2015). Similar phylogenetic relationships in the genera of sciaenids were noted based on molecular analyses (Lo et al. Citation2015; this study). Therefore, more mitogenomes are needed to fully elucidate the evolution of O. ruber and the phylogenetic relationship of the sciaenid species.
Disclosure statement
This study was supported by the Oceans & Fisheries Bureau of Fuzhou (grant no. XDHT2016051A), and the National Natural Science Foundation of China (grant no. 41476149). The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chen X, Ai WM, Xiang D, Chen SB. 2014. Complete mitochondrial genome of the red stingray Dasyatis akajei (Myliobatiformes: Dasyatidae). Mitochondrial DNA. 25:37–38.
- Lo PC, Liu SH, Chao NL, Nunoo FKE, Mok HK, Chen WJ. 2015. A multi-gene dataset reveals a tropical New World origin and Early Miocene diversification of croakers (Perciformes: Sciaenidae). Mol Phylogenet Evol. 88:132–143.
- Nelson JS. 2006. Fishes of the world. 4th ed. Hoboken, NJ: John Wiley & Sons, Inc.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Sasaki K. 1989. Phylogeny of the Family Sciaenidae, with notes on its zoogeography (Teleostei, Perciformes). Mem Grad Sch Fish Sci Hokkaido Univ. 36:1–137.
- Sasaki K. 2001. Sciaenidae. In Carpenter KE, Niem VH, editors. FAO species identification guide for fishery purposes. The living marine resources of the Western Central Pacific. vol. 5: Bony Fishes Part 3 (Menidae to Pomacentridae). Rome: FAO; p. 3169.